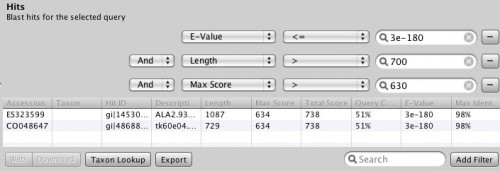
This is a mini-tutorial based on an example from “Gene und Stammbaeume” by Volker Knoop and Kai Mueller.
We will use Epos to create a Phylogeny of Marsupials based on the NAD5 Gene. The basic steps involved are: Collect initial data from the NCBI, search for homologue Sequences not covered by the initial search, find outgroup sequences, create an alignment and finally create a tree.
For this demo, we start with an empty Epos Workspace. If you want to create a new Workspace, use File->New Workspace…. and restart Epos.
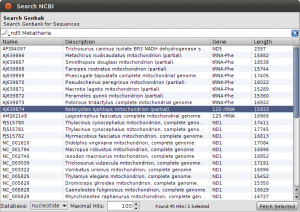
NCBI Search for nd5 Metatheria
To get things started, we have to search the NCBI for sequences linked to nad5 in Metatheria. You can do the search in the NCBI Web interface and download the results in GenBank format, but for now, we use the integrated NCBI Search in Epos. Select File->New->Search GenBank… and enter “nd5 Metatheria” into the search field before you hit enter. At the time of writing, I ended up with 45 complete mitochondrial genome hit. For ease of simplicity, select all the hits (STRG/CMD-A in the table), and press Fetch Selected. All selected hit will now be imported into your workspace, including the taxonomic information linked to the sequences.
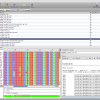
To see the imports and quickly check the taxonomy, select Sequences from the main toolbar. The sequence overview opens and you will see the 45 imported entries. Select all, and right-click on the table to get the context menu and choos Show Taxonomy… A window opens that shows the taxonomic hierarchy of the selected sequences entries. Verify that you have only sequences that are Metatheria.
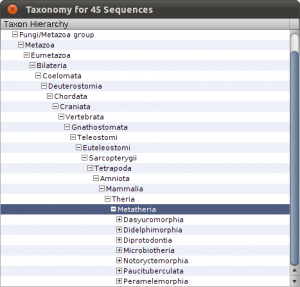
Check the taxonomy
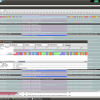
Right now, we have the complete genome sequences in the database, but we are interested only in the nd5 regions. We can use the Extract Features function, to get the DNA Sequences annotated as nd5. Again, select all sequences and hit the Extract Features button in the toolbar. The first dialog that appears asks you whether you want select from all features, or just the ones that are not unique. In our case, we want to see All Featrues but filter for CDS annotations. Type CDS into the filter field and hit Ok. Now we get a list of all CDS regions in all the selected sequences and you can check for the ones we are interested in. Use the search field in the lower right corner to filter for ND5 regions and click the checkboxes for the Features you want to extract. Before clicking Ok, make sure you checked the Keep Original Sequence Name box and Feature as Gene name. The first one ensures that the extracted sequences get the same name as the source sequence and the latter assigns a gene name to the extracted sequences.
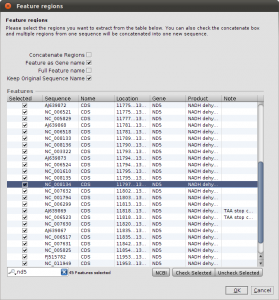
Extract ND5 Features
Back in the sequence view, we have 45 new Sequences, but the gene name column is a little bit messed up. Not all the annotations where named nd5. To fix this, select the new extracted sequences, right click and select Assign->Gene… and assign ND5 to all sequences. By default you find new data at the bottom of the list, so the last 45 sequences are the extracted features.
Before we continue creating alignments and trees, it is time to get a little bit more organized. To create collections of Data, you can use virtual files and folders in the epos workspace. To create a collection of our extracted sequences, right clock in the virtual file system browser on the left side of the epos main window (the blue area). Create a new Folder and name it ND5 DNA. Now select the extracted sequences and drag-and-drop them into the new folder. The folder icon becomes green and the folder contains your sequences. To view to content, right-click in the folder and select Show Content or use the little toolbar on top of the filesystem window. Note that you can also drag and drop folders into algorithms.
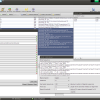
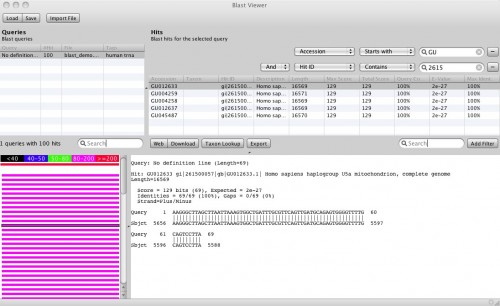
Okey, so before we create the first alignment, we want to extend our dataset further. The initial text based search might not have found all homologue sequences and we also need to find appropriate outgroups. We use Blast to extend our set of sequences. Select one of the nd5 sequences and hit the Blast button in the toolbar. This loads the sequence into the Blast dialog. We now want to do two blast searches. One for Prototheria and one for Eutheria to, find other homologues and outgroup sequences. Type Prototheria in the Taxon search bar to limit the search and add Prototheria to the Result name field before you click the submit button. When the job was successfully submitted, click on Show Jobs in the dialog that appears. This will show you the list of currently submitted jobs. Before you close the Blast search window, quickly submit another search. Set the taxonomic limit to Eutheria and resubmit the job.
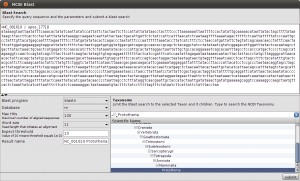
Blast Query
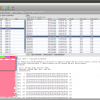
When both jobs are in the Done state, select them and click on Fetch Results in the Jobs window. Epos will now download the blast results and import them into the workspace. Click on Blast Results in the main toolbar. You will see the two blast datasets. Lets start with the homologue hits. Double click the results for the Prototheria search and confirm the next dialog with Ok, you do not have to customize the blast result import for this example. The result view opens and you should see a few hits for the query sequence. I found:
Tachyglossus aculeatus complete mitochondrial genome
Zaglossus bruijni complete mitochondrial genome
Ornithorhynchus anatinus mitochondrial DNA, complete genome
Select the hits and click the Download button in the lower toolbar. This will fetch the associated records from GenBank. When Epos asks you what to do with the downloaded results, click on Import to import them into the current workspace. As the hits are also complete genome sequences, you have to repeat the feature extraction steps. Select the sequences, choose extract features in the toolbar, filter for CDS, search for the nadh5 features. Note that the hits might not be well annotated. I found the three ND5 regions by filtering the table for “5” and checking the “Description” column. You can also hide table columns by right clicking the table header and remove the columns you are not interested in. Enable Keep the original sequence name and extract. Now assign ND5 gene names to all the newly extracted sequences and drag and drop the extraction to the ND5 DNA folder). You can switch to the sequence view using the Sequences button in the main toolbar.
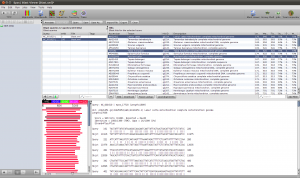
Blast Results for the Outgroup Selection
To select a few Outgroup sequences, open the results for the blast search restricted to Eutheria. There is a good chance you will not have hit the same sequences as this example. Just randomly picked three sequences. These are our Outgroup sequences.
Lemur catta
Talpa europaea
Mus terricolor
Again, these are all complete mitochondrial genomes, so we do the extraction step again. After you extracted the CDS regions and droped them into the ND5 DNA Folder, we can create the first Alignment and an initial tree.
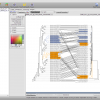
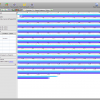
To create the alignment, open the ND5 DNA Folder content by selecting the folder and clicking the small sequence button in the toolbar on top or right click the folder and select Show Content->Sequences. You will see the content of the folder. Select all the sequences, right click and select Run. You will see a list of algorithms that can be applied to a set of sequences. Because we want to compute a multiple sequence alignment, select one of the alignment methods. For a fast alignment I choose Mafft. All the sequences are set as input and you can keep the parameters at their default values for now. Epos will warn you if Mafft is not yet installed. If that is the case, click on the Install Tool Button and use the Auto-Installation feature for mafft. Now the Run button on the bottom right of the toolbar should be enabled and you can click it to start the computation. Open the Jobs windows and wait until the job state is “Done” before you double click the job to fetch the results.
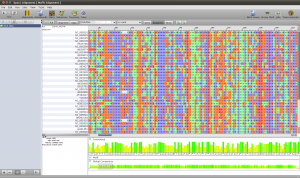
ND5 DNA Alignment
After the results are fetched and imported into the workspace, you can choose the Alignments view to get a list of all available alignments. The one we just computed should occur at the bottom of the list. For now, we use the Alignment as is and just right click it and select Run again to compute a tree.
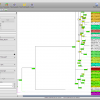
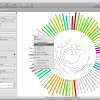
To get a quick result, choose NJ Tree to compute a neighbor joining tree from the alignment. When the NJ window opens, set the rooting method to “Outgroup” and select one of your outgroup sequences. You can go back to the sequence view to check for the right name. Now start the computation, it should just take a few seconds. Fetch the results and got to the Trees view using the main toolbar. Like all overviews in Epos the Trees view shows a list of all the trees in the current workspace and the computed NJ tree will be last in the list. Double click the tree to go the the detail view.
Note that the tree now shows the sequence names as labels. However, to get a better idea of what is where in the tree you can switch to a taxonomic labeling. Because you imported all your sequences from GenBank, they all come with taxonomic information, which is also imported into the Epos workspace. On the left side of the tree view, in the Nodes tab, turn on Taxon Names and the taxonomic naming will be used.
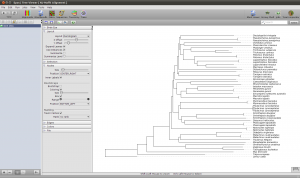
The first ND5 DNA Tree