About EPoS
EPoS is a modular Software Framework for Phylogenetic analysis with a focus on algorithms, computational methods, data management, visualization, and usability. The core modules bundled with EPoS can manage all the basic data types used to create phylogenetic trees and store your data in the EPoS workspace.
Sequences – You can import and export sequences from and to raw sequence files in different formats (fasta, genbank, embl, phylip, nexus,…) and you can fetch data from the NCBI using GeneId or Accession numbers.
Alignments – You can import and export sequence alignments from files or create alignments directly in epos. Currently we support the following alignment algorithms:
- ClustalW
- Mafft
- Muscle
- TCoffee/MCoffee
- Exact and Divide and Conquer (DCA/MSA)
- Dialign-TX
and Profile Alignments with:
- TCoffee
- ClustalW
Trees – Alongside direct file import, you can create phylogenetic trees with one of the following methods directly from alignments:
- Neighbor Joining
- Agglomerative Clustering
- RaxML
- MrBayes
- Paup Maximum Parsimony
Bot, RaxML and MrBayes support multi gene analysis, and so does the EPoS Framework. You can merge Alignments or manually annotate your alignment and than execute RaxML or MrBayes on on the annotated/merged Alignment.
Epos also supports methods to create Consensus and Supertrees directly from within the Framework. Supported methods are:
- N-Consensus (i.e. Strict or Majority Rule)
- Adams Consensus
- Maximum Agreement Subtree (base on Paup)
- Build
- Ancenstral Build
- MinCut and Modified MinCut Supertree
- FlipSupertree
- Supertree by Matrix Representation with Parsimony (Paup based)
- Physic and PhySIC IST
- Build with Distances
To analyse the resulting trees, we provide a sophisticated tree comparator, that can be used to compute distances between trees as well as a pairwise visual comparison.
Distance Matrices – You can import distance matrices or create a matrix from an alignment
Taxonomy – The EPoS Taxonomy module allows you to create and manage a custom Taxonomy, or import Taxa from NCBI. Sequences and Tree nodes can than be linked to Taxa to create an alternative naming scheme while keeping the original names. For example, with links to taxa, you can view your tree with different Taxonomic ranks (Species, Genus,…) and compare trees directly, based on the taxonomy, instead of manually editing the node labels.
Blast – The Epos Core provides access to the NCBI Blast WebService as well as local blast installations. This allow you to blast sequences directly against the NCBI or create your own Blast Database from a set of sequences. You can then import the results and brows the blast hits, while importing interesting hits directly into to Epos workspace. Beside the import of blast hits, you can also quickly annotate your Query Sequences based on the Annotations of the hit sequences.
- Genbank Import
Import Sequences from GenBank using an Accession number, GI Number or the link from the NCBI web page
- Feature Extraction
The Feature Extraction Dialog allows to extract annotated regions from sequences
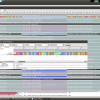
- Sequence Overview
The Seqence Overview that shows all sequences in the current workspace
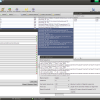
- Sequence View
The Epos Sequence View showing an annotated Sequence
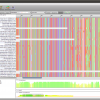
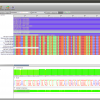
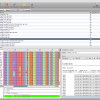
- Alignment View
Epos Alignment view with Taxonomic Naming
- Alignment Annotations
Alignment view that shows Sequence annotations
- Quick Align
Quick Pairwise comparison
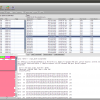
- Blast Results
View your blast results and import interesting hits into the workspace
- Sequence annotator
Use the blast hits to annotate your sequences
- RaxML multi gene configuration
- RaxML Configuration
- Alignment with annotations
- Alignment with annotations
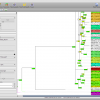
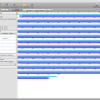
- Bootstrap Values
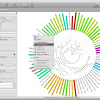
- Circular Tree
- Alignment Overview
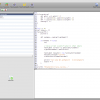
- Script support
Groovy script editor
- Epos Blast Dialog and Tool Installation
Epos supports automatic installation for most of the external tools.
- Annotated Sequence
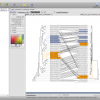
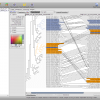
- Tree Comparison
- Tree Comparison
- Edge Compression
Epos can compress tree edges based on their weight
- Tree Overview