Tutorial: Merging Alignments and RaxML Multi-Gene analysis
In this tutorial we will create two alignments and perform a multi-gene analysis using RaxML. To get things started lets fetch data from the NCBI. I randomly picked 5 Taxa from “A multigene phylogeny of the Dothideomycetes using four nuclear loci” for the SSU and LSU regions.
Start Epos and go to File->New->Fetch from GenBank. Add the following Accession numbers for SSU:
AY584667,DQ678000,DQ678012,DQ471039,DQ678043
and for LSU:
AY584643,DQ678053,DQ678064,DQ470987,DQ678097
(Note that you can copy paste the whole line, Epos will split by comma). Hit Add and then Fetch the sequences. You will end up with 10 new sequences in your workspace. Click on the Sequences button in the main toolbar and select the SSU sequences (Epos keeps the order, so the first five should be SSU while the last five are LSU, but you can also check the description of the sequences. You can also select the sequences, right-click and select Assign->Gene… to assign SSU or LSU as gene names). With the SSU sequences selected, click the Run button in the toolbar or right click and select Run. Select ClustalW (or any other alignment method). The ClustalW configuration appears. If ClustalW is not installed on you machine, click on Install Tool to specify the path to ClustalW or Auto-install. Set the result name to “SSU” and keep the rest of the algorithm configuration. Now hit the Run button in the lower right corner. A job will be started on your local machine.
Repeat the steps for LSU. Select the LSU Sequences, click on Run and select ClustalW, set the result name to LSU and start the algorithm. Now show the Jobs manager by clicking on Jobs in the main toolbar. You should see your two ClustalW runs and they are probably already in the Done state. If not, wait until both jobs are “Done“. Now select the two jobs and press “Fetch” to load the results into your workspace. Click the Alignments button in the main toolbar and you will see a list of all alignment in your current workspace including the two ClustalW alignments for LSU and SSU.
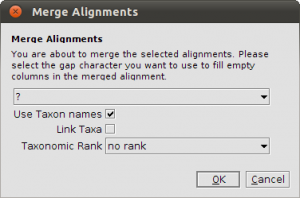
Now comes the tricky part. We have to merge the alignments to perform a multi-gene analysis. Select both alignments and click the Merge button in the toolbar. In the dialog that appears you have to check the Use Taxon names box! Because we fetched the sequences from GenBank, the Accession number is used as sequence name and the Accession numbers, of course, are not matching up. For example, AY584667 is the SSU and AY584643 is the LSU. They should appear as one row in the merged alignment. To avoid heavy renaming, we can use the taxonomic information to create a proper merge. With the sequence information, we also fetched the taxonomic information from the NCBI Taxonomy Database. Now both AY584667 and AY584643 are linked to the same taxon: Acarosporina microspora. Checking Use Taxon Names force Epos to merge based on the taxonomic information instead of the raw sequence names. Click in Okey and Epos creates the merged alignment, which will appear in the list of alignments in your workspace.
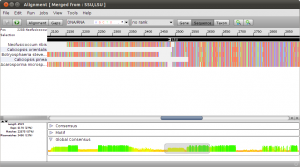
To get an idea of what happend, double click the new merged alignment. Notice the black arrows above the alignment. These are annotations on the alignment that mark different regions. In this example, the annotations represent the SSU and LSU regions in the merged alignment.
To perform a multi-gene analysis, use the Run button in the toolbar of the alignment view. You will see a list of algorithms that work on alignments. Select RAxML BS and ML to perform a rapid RaxML analysis with bootstrapping. If RaxML is not yet installed, use the Install Tool button to install it. Select a model, for example, GTRCAT and you are good to go. To verify that you are doing a multi-gene analysis, click on the small right-arrow button above the RaxML configuration parameters. You will see a table of regions used for the analysis. Here you can also choose different models for the regions if you wish. Epos automatically uses all available alignment annotations as regions for RaxML. To start the algorithm, click the Run button in the lower right corner and open the Jobs window after successful submission. When the RaxML run is finished and in Done state, double click the job to fetch the results. Click on Trees in the main toolbar to view all the trees in your workspace. The RaxML tree is also in the list.
Foursquare as forefront money spinner for geolocation services |
Coenzyme Q10 (CoQ10) is a substance similar to a vitamin. It is found in every cell of the body. Your body makes CoQ10, and your cells use it to produce energy your body needs for cell growth and maintenance. It also functions as an antioxidant, which protects the body from damage caused by harmful molecules. CoQ10 is naturally present in small amounts in a wide variety of foods, but levels are particularly high in organ meats such as heart, liver, and kidney, as well as beef, soy oil, sardines, mackerel, and peanuts. .
Our blog
<.http://www.foodsupplementcenter.com/best-fiber-supplement/
銅材
diablo 3 Lord of expert knowledge greedy…….. RIP north blizzard…… burn in hell fashionable greed blizzard
I called into stereo this morning because these folks were saying, “Wow a lot of folks are playing Wow. Must be a tasty game, not just for nerds. ” Having had 3 accounts previously I contacted to inform them they will weren’t reading. Half of those subscriptions have been in Asia. Why? Because of the a lot websites that offer gold and in portions of 5, 000+ for each and every server, people don’t understand we now have literal sweat shops where kids are made to farm for gold. If you played warcraft and realize how hard it truly is to make gold, how can every service provider offer 5, 000+ gold for each and every server out there? Simple Mass amounts of accounts useful for farming gold. So the numbers are inflated I reported and yes, the game is pleasure, but when you factor during the time of you have to part with kissing guild butt exactly how guilds run your lives and try to raid (god forbid you have life and miss a raid) and then no longer becomes gaming but a job. I’m sorry I already used. I don’t need another job which i’ve to pay for. My third account We a lvl 70 holy priest, pretty high tier items, and then one morning, 800-900g and said, “I’m done. ” And I will not play again. Ever. So I deleted her or him. From head to toe across all my guildies as well as can said, “Goodbye. ” My friends on the market me $100 for my personal account. I hated WoW plenty of I said sorry and threw the software away apart, and then celebrated. I went back to living gaming. This news serves in order to buffer the truth. All my friends feel canceled their accounts. Game reviewers have been complaining available online low population on secured once thriving servers. If Wow just started using it through their heads to provide decapitate guild heads (teeny HOA’s in my experience), make the game playable with single-player however rented in game shield, such as earn gold and hire your matter, then everything would be good, because as it cases now, everything in WoW will depend on one thing Your guild. And every guild available sucks. Why? Abuse of power, spamming, stabbing each other in the back over virtual something, and just plain dumb inane and incessant chat amounting to crap. Chuck Norris? Need I say accessories. What was fun once is infuriatingly frustrating to put it mildly. Why put such a great game to guild masters who are nothing more than geeks with no lifestyle. Sure, there are good guilds. But these guilds are far and few between and include things like friends who can look there windows of where they live and see having to not take the game so seriously you to definitely become Hugo Chavez. Good luck to all you could Wow fanboys that must pay back seriously creeped me out by being if my character was part of their daily lives. Scary.
hola wordpress administrator, I can see that you’re making use of wordpress. I would like to tell you about a exceptional member site from which it is possible to get lots premium themes and plugins.If you decide to become a member please use my affiliate link.http://themesandplugins.co.uk?ref=shirley
It’s hard to find your posts in google. I found it on 21 spot, you should build quality backlinks ,
it will help you to get more visitors. I know how to help you, just
search in google – k2 seo tips
I read a lot of interesting content here. Probably you
spend a lot of time writing, i know how to save you a lot of work,
there is an online tool that creates unique, SEO friendly articles in minutes, just type in google – laranitas free content source
Turnierkleider
Epos
Tauchschule Hurghada
Epos
ทางเข้า 918kiss
Epos
ดูหนังฟรี
Epos
http://www.happe-druck.de
Epos
deutschebewertungen.de
Epos
tauchbasis hurghada
Epos
Monitorwerbung
Epos
Zauberer Schweiz
Epos
Businessportraits München
Epos
paintball outdoor
Epos
Robert Franz Produkte
Epos
Quercetin Studien
Epos
stores4fashion.com
Epos
Hotel
Epos
Bio Suppen kaufen
Epos
autouncle.de
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
luxury-chalet-kreischberg.com
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
planet-soft.de
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
ratingen-webservices.de
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
Strip Clubs in Germany
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
online dating success
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
organicpack.de
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
escortfor.me
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
SEO Texte kaufen
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
juicerystore.com
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
Premium Nahrungsergänzung
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
LED Buchstaben
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
exittheroom.de
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
Zauberer Basel
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
CBD Öl 38% kaufen
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
Fluch aufheben
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
Feuerwehr Streetwear
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
paketversand24.com
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
hairdoshop.de
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
Kopftuch
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
Anonyme Wegwerf-EMail Adresse
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
Rohrreinigung Berlin
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
neon
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
fix-text.de
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
ateliersv.de
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis
relocation
Epos » Blog Archive » Tutorial: Merging Alignments and RaxML Multi-Gene analysis