Every (put a large number here) days I stumble over the question, “how large is the universe of small molecules?” in a publication or a blog. Not the number of small molecules that we already “know” – those are covered in databases such as HMDB, PubChem or ZINC. (Although many compounds in PubChem etc. are purely hypothetical, too.) Not the number of metabolites that actually exist in nature. Also, we do not care if these small molecules are synthesizable with current technology. Rather, we want to know: How large is the space of small molecules that could exist? These could be natural small molecules (natural products, secondary metabolites) or synthetic compounds. I usually do not like the numbers that people report as “the truth”, as one estimate was taken out of context and started to develop a life of its own. Hence, I thought I write down what I know.
Obviously, we have to first define precisely what we actually counting. Since the problem is already hard enough, we usually do not care about 3d considerations (usually including stereochemistry), and simply ask for the number of molecular graphs (chemical 2d structures). On the one hand, resulting estimates are a lower bound, as adding stereochemistry into the mix results in more structures; on the other hand, resulting estimates are upper bounds, in the sense that many molecular graphs that we draw, cannot exist in 3d space. For the sake of simplicity, let’s forget about this problem.
Next, we have to define what a “small molecule” actually is. Usually, we simply apply a threshold on the mass of the molecule, such as “all molecules below 1000 Dalton“. If you prefer, you may instead think of “molecular weight”; this is the expected mass of the molecule, taken over all isotopologues and, hence, slightly larger. Yet, people usually consider mass in these considerations. Maybe, because mass spectrometry is one of the (maybe, the) most important experimental technology for the analysis of small molecules; maybe, because isotopes make a big difference for small molecules. Since our bounds on mass are somewhat arbitrary, anyways, this is not a big problem: Why exactly 1000 Dalton? Is prymnesin-B1 a small molecules? It is not a peptide, not a lipid, not a macromolecule, and it sure looks like a biological small molecule; it is just “too heavy” (1819.5 Dalton). For our calculations we will stick with the mass of 1000 Dalton; yet, it all still applies when you want to use a different threshold, and doing so is actually very simple, see below.
Now, the number of small molecules that people are reporting and repeating, making this number “more and more true” (to the point that Google Gemini will return that number as the correct answer when you query with Google) comes from a paper by Bohacek et al. (1997). The caption of Figure 6 contains the sentence “A subset of molecules containing up to 30 C, N, O, and S atoms may have more than 1060 members”. The authors make some back-of-the-envelope calculations in a long footnote, to further support this claim. The authors clearly state that the actual numbers they are giving are incorrect, for each step of their calculations. These calculations are merely meant to demonstrate that the true number is enormously huge; and, that the actual number is not that important, and might never be known to mankind. Unfortunately, many readers did not grasp the second part of what the authors were saying and instead, starting reporting the number “1060 small molecules” as if this number had been calculated for good. From there, it was only a small step into a Science/Nature news (I have to search for it), publications, websites, and finally, the “knowledge” of a large language model such as Gemini. The Wikipedia article about Chemical Space is much more cautious, and also mentions the 500 Dalton cutoff that Bohacek et al. assumed, as they were interested in pharmacologically active molecules. Funnily, Google Gemini does not link to the Wikipedia article, but rather to a “random” website that cites the number of Bohacek et al..
Let us see if we can do better than that. First, we look at a few things that are known. The On-Line Encyclopedia of Integer Sequences (OEIS) gives us loads of helpful numbers:
- Alkanes are hydrocarbons (carbohydrates with no oxygen) with the “minimal” molecular formula Cn H2n+2 and have no multiple bonds, a tree structure, and no cycles. The number of alkanes can be found in sequence A000628, see also here. Yet, this sequence counts each individual stereoisomer, different from what we said above. The sequence A000602 counts different stereoisomers as identical. For upper bound 1000 Dalton we use n = 71 (nominal mass 996 Dalton), and this is 1 281 151 315 764 638 215 613 845 510 structures, about 1.28 · 1027, or 1.28 octillion. Clearly, not all of them are possible; yet, the number of of “impossible” structures may be smaller than one may think. Alkanes are small molecules, so this is a lower bound on the number of small molecules of this mass.
- In fact, we can approximate – in the colloquial and in the mathematical sense – this number using the closed formula (i.e. the formula has no recurrences)
0.6563186958 · n-5/2 · 2.815460033n,
see Table 3 and equation (16) here (reprint here). For n = 71 this approximation is 1.279 · 1027, which is already pretty close to the true (exact) number. Importantly, this allows us to classify the growth as exponential, meaning that increasing n by one, we have to multiply the number of molecules by a constant (roughly 2.8). We can safely ignore the non-exponential part of the equation, as its influence will get smaller and smaller when n increases. Exponential growth might appear frightening, and in most application domains it would be our enemy; but for counting small molecules, it is actually our friend, as we will see next. - Molecules are no trees, they have cycles. So, another idea to get an idea on the number of small molecules is to count simple, connected graphs. “Simple” means that we do not allow multiple edges between two nodes (vertices). We have different types of bounds (single, double, etc.) but multi-edges means that there can be 1000 edges between two nodes, and that is definitely not what we want. We also ignore that we have different elements, that is, labeled nodes. This number is found in sequence A001349. This number growths super-exponential, meaning that it growths faster (for n to infinity) than any exponential function. That is fast, and we indeed observe that already for reasonable number of nodes, numbers become huge: For n = 19 nodes it is 645 465 483 198 722 799 426 731 128 794 502 283 004 (6.45 · 1038) simple graphs; for n = 49 we already have 1.69 · 10291 simple graphs. That is a lot, compare to the number of alkanes. Even more intimidating is the growth: Going from n = 49 to n = 50 nodes we reach 1.89 x 10304 simple graphs, a factor of 11 258 999 068 424 (11 trillion) for a single node added. Suddenly, factor 2.8 does not look that bad any more, does it?
- Now, one can easily argue that simple graphs are too general: The number of edges incident to (touching) a node is not restricted, whereas it is clearly restricted for molecular graphs. For simplicity, let us consider Carbon-only molecules; carbon has valence 4. Let us further simplify things by only considering cases where each node is connected to exactly 4 other nodes (chemically speaking, no hydrogen atoms, no double bonds). The number of 4-regular, simple, connected graphs is sequence A006820. Given that there is a single such graph for n = 6, it might be surprising how quickly numbers are increasing even for moderate number of nodes n: For n = 28 there are 567 437 240 683 788 292 989 (5.67 · 1020) such graphs.
All of these numbers give us a first impression of the number of small molecules we could consider. Yet, none of them give us any precise number to work with: The number of alkanes is just a lower bound, the number of simple graphs is too large, and the number of 4-regular simple graphs is simultaneously too large (we cannot simply demand that two Carbon atoms are connected, we somehow have to realize that in 3d) and too small (4-regular, Carbon-only). That is not very satisfactory. Yet, we can already learn a few important things:
- The coefficient c before the 10k is actually not very important. If we are dealing with numbers as large as 10100, then a mere factor of below ten does not make a substantial difference. It is the exponent that is relevant. Similarly, it does not make a relevant change if we think about mass or nominal mass.
- If we have exponential growth, as we did for alkanes, then it is straightforward to calculate your own estimate in case you are unhappy with the upper bound on mass of 1000 Dalton. Say, you prefer 500 Dalton; then, the estimate for the number of alkanes goes from 1027 to 1027/2 = 3.16 · 1013. Similarly, for 1500 Dalton, we reach 1027·1.5 = 3.16 · 1040 alkanes.
Yet, we can also look at the problem empirically: There exist different methods (the commercial MOLGEN, OMG, MAYGEN, and currently the fastest-tool-in-town, Surge) to generate molecular structures for a given molecular formula. For example, Ruddigkeit et al. generated “all” small molecule structures with up to 17 heavy atoms (CNOS and halogens). I put the “all” in apostrophes because the authors used highly aggressive filters (ad hoc rules on, does a molecule make sense chemically? etc.) to keep the numbers small. With that, they generated the GDB-17 database containing 166 443 860 262 (1.66 · 1011) molecular structures. Unfortunately, only about 0.06% of that databases are publicly available. Also, the aggressive filtering is diametral to our question, how many molecular structures exit.
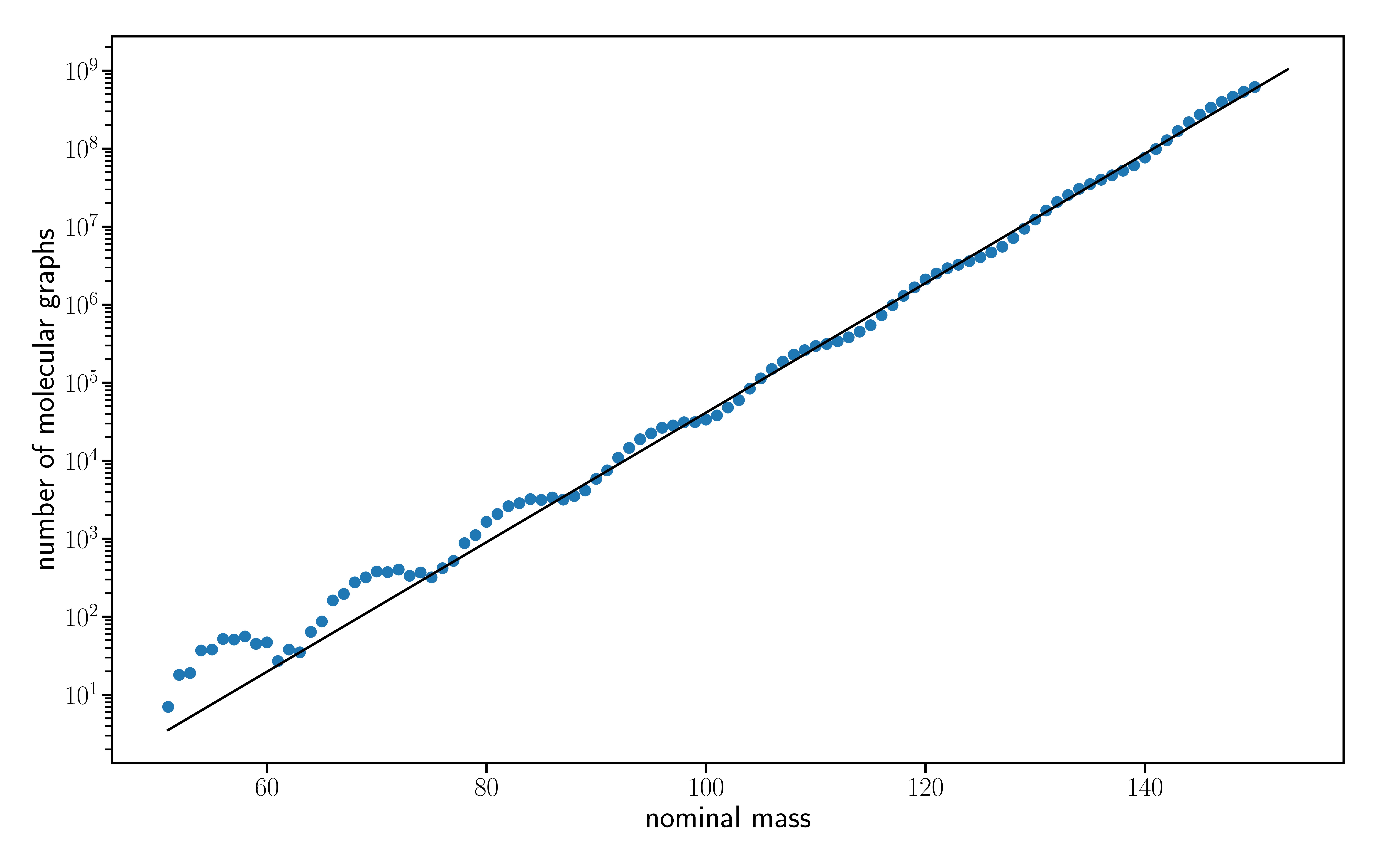
More interesting for us is a paper from 2005: That year, Kerber et al. published the paper “Molecules in silico: potential versus known organic compounds“. For the paper, the authors generated all small molecule structures with nominal mass up to 150 Dalton, over the set of elements CHNO, using MOLGEN. For example, MOLGEN constructs 615 977 591 molecular structures for mass 150 Dalton. This paper also contains the highly informative Figure 1, where the authors plot the number of (theoretical) molecular structure against nominal mass m. Looking at the plot, I could not resist to see exponential growth, which (if you use a logarithmic y-axis) comes out as a simple line. Remember that exponential growth is our friend.
So, let’s do that formally: I fitted a regression line to the empirical numbers (see Chapter 10 here), and what comes out of it is that we can approximate the number of small molecules between 80 and 150 Dalton as 2.0668257909 · 10−4 · 1.2106212044m. Be warned that there is not the slightest guarantee that growth will continue like that for masses larger than 150 Dalton; it is highly unlikely that growth might slow down, but given that the number of simple graphs is growing superexponentially, maybe the same is true here? I cannot tell you, I am not that much into counting and enumerating graphs. But in any case, my Milchmädchenrechnung should make a much better estimate than anything we have done so far; to be precise, much better than anything else I have ever heard of. For nominal mass 1000 Dalton this number is 2.11 · 1079 molecular structures.
But wait! The above formula tells us the (very approximate) number of all small molecules with mass exactly 1000 Dalton; what we want to know is the number of molecules with mass up to 1000 Dalton. Fun fact about exponential growth: These numbers are basically the same, with a small factor. Recall that the number of molecules with mass m equals a · xm. Pretty much every math student has to prove, during his first year, the equation
To this end, if we want to know the number of molecules with mass up to 1000 Dalton, we simply use the number of molecules with mass exactly 1000 Dalton, and apply a small correction. In detail, the number of molecules with mass up to 1000 Dalton is (almost exactly) 1.21062 / (1.21062 – 1) = 5.74786 times the number of molecules with mass exactly 1000 Dalton. We reach an estimate of 1.21 · 1080 small molecules over elements CHNO with mass up to 1000 Dalton. For mass up to 500 Dalton it is about 1040 molecules (which tells us that the inital estimate was rather far off, and I do not expect that even a superexponential growth could correct that); for mass up to 1500 Dalton it is 10120 small molecules.
But, alas, this estimate is for elements CHNO only; what if we add more elements to the mix? For halogens it is mainly fluorine that will substantially increase the number of molecular structures: Halogens are much heavier than hydrogen. If we replace a single H by F in a molecular structure, then the nominal mass of the molecule increases by 8. The above estimate tells us that there are only 1/4.61 = 0.216 times the molecular structures to consider if we decrease the mass by 8. On the “plus side” we can replace any H by F to reach a different molecule, if we gallantly ignore the problem of isomorphic graph structures. For more than one fluorine, the combinatorics of where we can add them growths polynomially; but at the same time, the number of structures we start from decreases exponentially. Hence, one should not expect a substantially higher number if we add fluorine to the mix; and an even smaller effect for the other halogens. More worrying, from a combinatorial standpoint, are the elements sulfur and phosporus. In particular sulfur with valence 6 could wreak havoc of our estimates. I don’t know what happens if you allow an arbitrary number of S and P; I also don’t know if we would even want such all-sulfur molecules. The only number that I can report is again from the Kerber et al. paper: They also generated all molecular structures for nominal mass 150 Dalton and elements CHNOSiPSFClBrI, using the (unrealistic) fixed valences 3 for phosphorus and 2 for sulfur, resulting in 1 052 647 246 structures, compared to 615 977 591 molecular structures over elements CHNO. Yet, this small increase in structures (merely doubled, phew) should not lull us into a false sense of security: Not only the unrealistic valences, even more so the relatively small mass could mean that in truth, the number of structures with sulfur and phosphorus is substantially higher. I suggest we stick with the number 1080 and in the back of our minds, keep the note “it is likely even worse”.
It is not easy to grasp a number as large as 1080, so a few words to get a feeling:
- The number 1080 is 100 septemvigintillion, or 100 million billion billion billion billion billion billion billion billions. If we have a name for it, it cannot be that bad.
- The observable universe contains an estimated 1080 atoms. If we had the technology to build a computer that uses every single atom of the observable universe as its memory, and where we need only a single atom to store the complete structure of a small molecule, then the observable universe would be “just right” to store all small molecules up to 1000 Dalton.
- Using the same technology, molecules up to 1010 Dalton would be absolutely impossible to store, and 1100 Dalton would be absurd. For molecules up to 2000 Dalton you would need 1080 observable universes. Alternatively, a technology that puts a complete universe into a single atom, and does so for each and every atom of this universe. See the final scene of Men in Black.
- For comparison, the total amount of storage on planet Earth is, at the moment, something like 200 zettabytes, mostly on magnetic tapes. If we could store every molecular structure in 2 bytes, this would allow us to store 1023 molecular structures. If we could now again fold all the storage into a single byte, and again, and again, we reach – after four iterations of folding – the required memory.
- If we have a computer enumerating molecular structures at a rate of 1 billion structures per second, then we could enumerate 3.15 · 1016 structures per year, and it would take 3.17 · 1063 years to enumerate them all. For comparison, the estimated age of the universe is 1.38 · 1010 years. We would need more than 1050 universes to finish our calculations, where in each universe we have to start our computations with the big bang.
- Maybe, it helps if we use all computers that exist on planet Earth? Not really; there are maybe 10 billion computers (including smartphones) on Earth, but let’s make it 1000 billion, to be on the safe side. The required time to enumerate all structures drops to 3.17 · 1051 years.
- But maybe, we can use a quantum computer? Grover’s algorithm brings down the running times to
if we have to consider n objects. Again assuming that our quantum computer could do 1 billion such computations per second, we would end up with a running time of 1031 seconds, or 3.17 · 1023 years. That is still a looong time. There might be smarter ways of processing all structures on a quantum computer than using Grover’s algorithm; but so far, few such algorithms for even fewer problems have been found.
Now, 1080 is a large number, and you might thing, “this is the reason why dealing with small molecules is so complicated!” But that is a misconception. It is trivial to find much larger numbers that a computer scientist has to deal with every day; just a few examples from bioinformatics: The number of protein sequences of length 100 is 1.26 · 10130. Yet, we can easily work with longer protein sequences. The number of pairwise alignments for two sequences of length 1000 is 2.07 · 10600. Yet, every bioinformatics student learns during his/her first year how to find the optimal alignment between two sequences, sequences that may be much longer than 1000 bases. The thing is, sequences/strings are simple structures, and we have learned to look only for the interesting, i.e. optimal alignments. (Discrete optimization is great!) What makes dealing with small molecule structures complicated is that these structures are themselves rather complicated (graphs, when even trees can be a horror, ask someone from phylogenetics) plus at the same time, there are so many, which prevents us from simply enumerating them all, then doing the complicated stuff for every structure.
That’s all, folks! Hope it helps.