CANOPUS is a tool for predicting compound classes directly from MS/MS. Because it does not depend on any database, CANOPUS is suitable for the classification of unknowns. CANOPUS runs as a webservice and is integrated in SIRIUS from version 4.4.
Installation
Please, download the appropriate SIRIUS version for your operating system, here. You need version 4.4 or higher.
The GUI version (which also includes the full command line version) requires no external JRE. Everything ist included. Download, extract, execute.
Usage
See the SIRIUS documentation. Also see the CANOPUS Behind the scenes talk for method details.
Demo data
Download the demo data here and extract it.
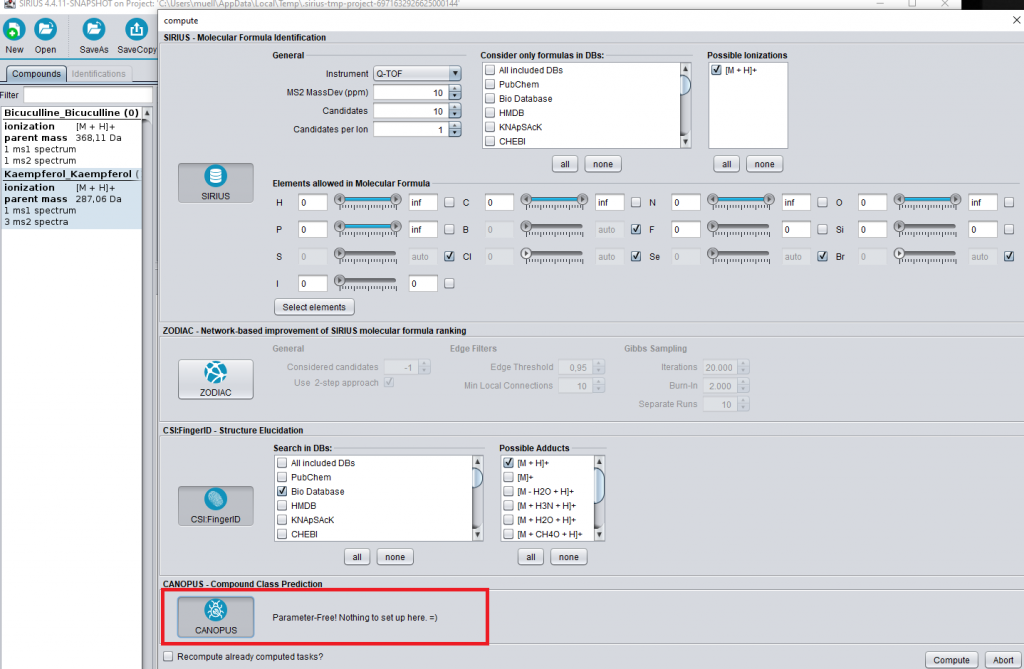
Drag and Drop the ms files in the ms folder into the SIRIUS GUI. Next, click on the “Compute all” button in the toolbar.
Check the buttons for “SIRIUS”, “CSI:FingerID”, and “CANOPUS”. When analyzing biological samples, we recommend to also enable ZODIAC.
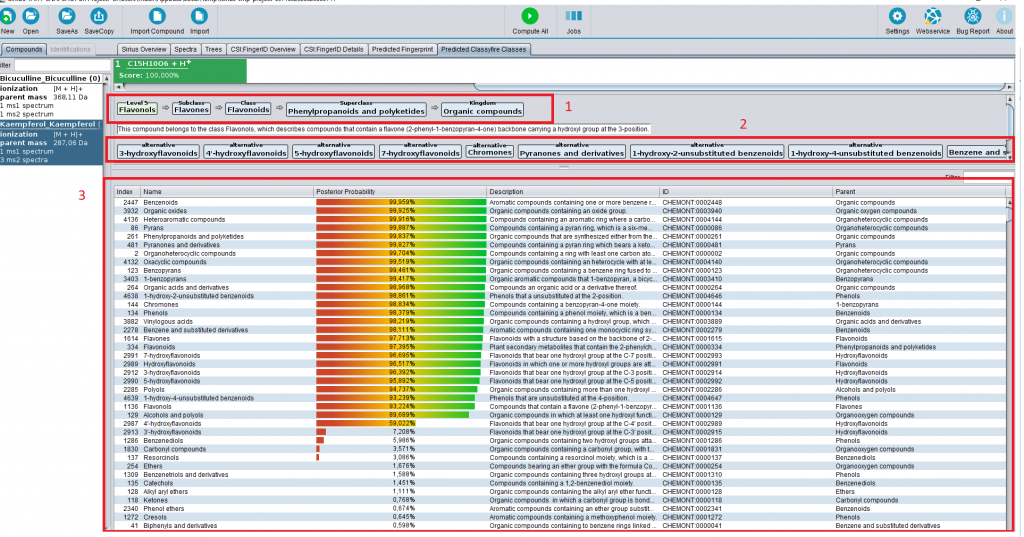
Click on compute and wait while the data are processed (which should take only a few seconds). The CANOPUS classification will appear in the “Predicted ClassyFire classes” tab.
(1) The most informative class (light green), and its ancestor classes (light blue).
(2) Alternative classes. In the ClassyFire chemontology, every compound is assigned to multiple classes. In this example, the compound kaempferol is a flavonoid, but also a benzenoid.
(3) The table lists all ClassyFire classes, with description parent class and so on. The colored bar denotes the predicted probability for this class. Only classes with probability above 0.5 are listed in (1) and (2).
Jupyter notebook for analyzing large datasets with CANOPUS
We provide a python library for summarizing and visualizing CANOPUS results: https://github.com/kaibioinfo/canopus_treemap
We recommend to use this python library within a jupyter notebook. An example notebook can be found here. This notebook is also a tutorial, demonstrating how CANOPUS annotations can be used to analyze large datasets.
References
K. Dührkop, L.-F. Nothias, M. Fleischauer, R. Reher, M. Ludwig, M. A. Hoffmann, D. Petras, W. H. Gerwick, J. Rousu, P. C. Dorrestein, and S. Böcker. Nat Biotechnol, 2020. https://doi.org/10.1038/s41587-020-0740-8