Gecko3 is an open-source software for finding gene clusters in 500 and more genomes on a laptop computer. The underlying gene cluster model can cope with low degrees of conservation and misannotations, and is complemented by a sound statistical evaluation. Gecko3 does not assume monophylecity or collinearity of gene clusters. Gecko3 offers an easy-to-use graphical user interface, but can also be used as a command line tool in an analysis pipeline.
Gecko3 is an open-source software for finding gene clusters in 500 and more genomes on a laptop computer. The underlying gene cluster model can cope with low degrees of conservation and misannotations, and is complemented by a sound statistical evaluation. Gecko3 does not assume monophylecity or collinearity of gene clusters. Gecko3 offers an easy-to-use graphical user interface, but can also be used as a command line tool in an analysis pipeline.
Installation
Gecko3 can be downloaded as a full bundle, including all dependencies. It requires Java 7 to run, see below. The source code can be found here.
| Version | Bundled | Sources |
| 1.0 | Gecko3.zip | |
| 1.1 | Gecko3.1.zip |
Gecko3 does not need to be installed. Download and extract the zip file to any folder (for example, your user directory or D:\ for Windows, or your Application folder for Mac OS X). This creates a new folder “Gecko3”. Open this folder: You find the file “readme.txt” that contains more information on installing and running Gecko3. Open the subfolder “bin”:
- If you are using Windows, the file “Gecko3.bat” is the right one for you. You might want to create a link to this file: Click and drag the file to the desktop, keeping the ALT key pressed. You can rename this link (the one on your desktop) as you like. You start Gecko3 by double-clicking this link.
- If you are using Linux, then the file “Gecko3” is your friend. Move to the “bin” folder and type “./Gecko3”.
- If you are using Mac OS X, it is again the file “Gecko3”. If you are not allowed to run the file, go to your System Preferences, select “Security & Privacy” and allow Mac OS X to run Gecko3. Then again open the file “Gecko3”. Mac OS X will probably ask you again whether you are sure you want to open it. Click open to start Gecko3. If you want to create an alias to start Gecko3 directly from your Desktop, right-click on the file, select “Make Alias”, and move it to your Desktop.
Gecko3 requires Java 7 to run. For Windows 7 and Mac OS X, Gecko3 can also be run using Java 8; we will report here as soon as we are sure that Gecko 3 and Java 8 cooperate for Linux, too. You can use Oracle Java or OpenJDK and may use either the JRE (Java Runtime Environment) or the JDK (Java Development Kit).
Gecko3 does not have minimal requirements beyond this, but to process a large dataset, you will need sufficient memory. We recommend 8GB RAM for analyzing 500+ bacterial genomes.
Java Installation
- Windows 64-Bit: Oracle Java JRE is recommended. Follow this link and select “Windows Offline (64-Bit)”. Download and execute the file to install Java JRE.
- Linux: OpenJDK 7 is recommended. How to install Java, depends on your specific Linux distribution. You should be able to install Java 7 or Java 8 from your package management system.
- Mac OS X ≥ 10.7.3: Oracle Java JRE is recommended. Follow this link and select “Mac OS X”. Download and execute the file to install Java.
Also see the FAQ at the end of this page.
Sample data
Download the following test data for Gecko3:
- Synechocystis.Default.zip: A large dataset of 678 genomes from RefSeq with homology families from the STRING database. Clusters are computed with the default distance table using Synechocystis sp. PCC 6803 as reference. We recommend 4GB RAM to view this dataset, 8GB if you want to re-compute clusters.
- 5Genomes.Default.zip: A small dataset of 5 genomes from RefSeq with homology families from the STRING database. Clusters are computed with the default distance table in all against all mode.
- 5Genomes.zip: A small dataset of 5 genomes from RefSeq with homology families from the STRING database, without precomputed clusters.
Usage
The following is a very short introduction of how to use the Gecko3 graphical user interface; a more detailed user guide can be found in the file “readme.txt” mentioned above.
For precomputed clusters, exported from Gecko3:
- Download one of the sample datasets (#1 or #2) provided above. We recommend not to store it in the same folder as the program files. Extract the zip file.
- Start Gecko3.
- Select File/Open, move to the folder where you stored the dataset, select it. This may take some time.
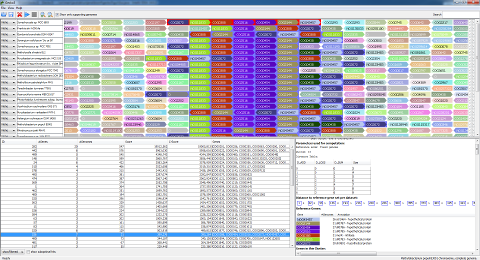
- Now, you see the Gecko3 results page. The detected clusters are in the lower left corner. The upper half contains a visualization of the genomes in the dataset. The lower right corner will contain information about the selected gene cluster, and is currently empty.
- Double-click on of the gene clusters on the lower left. You will see that the genome display changes, and that additional information is displayed in the lower right.
- By default, Gecko3 is now only displaying genomes the cluster appears in. To display all genomes, uncheck the “Show only supporting genomes” check-box above the genome display.
- The gene clusters look somewhat unordered. To improve the visualization, you can double-click on any gene in the gene cluster. This will align all homologous gene families in the detected gene clusters to this gene.
- If there is more than one occurrence of a gene cluster in a genome, you can use the arrows on the right to browse through the occurrences.
- You can force Gecko3 to display only those clusters where a particular genome has or has no occurrence. Use the buttons on the left of the genome names.
- By default, clusters are sorted by corrected p-value (C-Score, negative logarithm of the corrected p-value). Click on table headings in the cluster list to change this.
- By default, Gecko3 hides clusters with large overlap (showFiltered drop-down box below the list of clusters). You can change that to display all clusters.
- Right-click on a cluster and select “export” to export it as a PNG, JPG, or pdf file.
For a de-novo gene cluster computation:
- Download the sample datasets (3.) provided above. We recommend not to store it in the same folder as the program files. Extract the zip file.
- Start Gecko3.
- Select File/Open, move to the folder where you stored the dataset, select it.
- From the List of genomes, select all genomes you want to import. You can also click choose all at the bottom. Click OK to import the genomes.
- Now, you see the Gecko3 session with the genomes displayed in the upper half.
- Select the play symbol. You can choose either Single Distance or Distance Table and Click OK to start the computation with default parameters (default distance and size parameters, cluster has to be contained in all genomes, all genomes as reference).
- Note that computations are considerably longer if you use all genomes as references. For large datasets such as the Synechocystis sample dataset, you should instead choose one of the genomes as the reference. Select “Reference type: fixed genome”, then select one of the genomes.
- After the computations are finished, you see the Gecko3 results page, as in step 4 for the precomputed clusters.
- You can export the results by choosing File -> Save session and selecting a file you want to save the results to.
Publications
Algorithms
Katharina Jahn, Sascha Winter, Jens Stoye and Sebastian Böcker
Statistics for approximate gene clusters
BMC Bioinformatics, 14(Suppl 15):S14, 2013. Proc. of RECOMB Satelite Workshop on Comparative Genomics (RECOMB-CG 2013).
Katharina Jahn
Efficient computation of approximate gene clusters based on reference occurrences
J Comput Biol, 18(9): 1255-1274, September 2011. Proc. of RECOMB Satelite Workshop on Comparative Genomics (RECOMB-CG 2010).
Sebastian Böcker, Katharina Jahn, Julia Mixtacki and Jens Stoye
Computation of median gene clusters
J Comput Biol, 16(8):1085-1099, 2009.
Applications
Volker U. Schwartze, Sascha Winter, Ekaterina Shelest, Marina Marcet-Houben, Fabian Horn, Stefanie Wehner, Jörg Linde, Vito Valiante, Michael Sammeth, Konstantin Riege, Minou Nowrousian, Kerstin Kaerger, Ilse D. Jacobsen, Manja Marz, Axel A. Brakhage, Toni Gabaldón, Sebastian Böcker and Kerstin Voigt.
Gene expansion shapes genome architecture in the human pathogen Lichtheimia corymbifera: an evolutionary genomics analysis in the ancient terrestrial Mucorales (Mucoromycotina).
PLOS Genetics, 10(8):e1004496, 2014.
FAQ
- I am using Windows 32-Bit, can I also run Gecko3? Yes you can, but due to the restrictions of the Java Runtime Engine, you may not have enough memory to use Gecko3 for computing gene clusters in medium to large datasets. Visualization of the results might still be possible (gene clusters in 678 genomes use about 1.5GB RAM). Select “Windows Offline (32-Bit)” when installing Java. Select “Gecko3-32bit.bat” for running Gecko3.
- Can I use Oracle Java 8 available for Ubuntu? No, you should not use it, as it is known to cause display errors with Gecko3.
- Max OS X ships with Java, do I still have to install a Java version? Yes. Gecko3 does not work with Apple Java 6, the latests Java version OS X installed and updated by default. Later OS X version do not contain an Apple Java, but instead suggest installing Oracle Java.
- I am running Mac OS X with version before 10.7.3, how can I install Java 7? Java 7 installation is not possible for MAC OS X versions before 10.7.3. You need to upgrade to OS X 10.7.3 or higher.
- Is running Java a security problem? Only if you enable execution of Java programs in your web browser. Today, there is practically no need to run Java from the browser, so you should disable this option. In fact, Java became less of a risk in 2014. Always keep your JRE up-to-date.
- I am running Windows and I get an error message, “ERROR: JAVA_HOME is set to an invalid directory”. Probably, deleting the JAVA_HOME environment variable will solve the problem: follow instructions at the end of this page.
- I found a bug, what shall I do? You can directly create bug tickets in the Gecko3 Trac, either anonymous or after creating an account, so you will be informed about the status of your ticket. Please describe the bug as precisely as possible, to allow us to track it down.
Dependencies
| Library | Version | License | URL |
| args4j | 2.0.+ | MIT | args4j |
| colt | 1.2+ | Apache style | Colt |
| commons-math | 3.2 | Apache License 2.0 | Apache Commons Math |
| jgoodies-forms | 1.7.+ | BSD 2 | JGoodies FormLayout |
| glazedlists_java15 | 1.9.+ | LGPL 3.0 MPL 2.0 | Glazed Lists |
| itext | 4.2.1 | LGPL 3.0 MPL 2.0 | itext |
| guava | 18.0 | Apache License 2.0 | Guava |
| slf4j-api | 1.7.7 | MIT | SLF4J |
| logback-classic | 1.1.2 | EPL-1.0 LGPL-2.1 | Logback |
| junit | 4.+ | EPL-1.0 | Junit |
Authors
Sascha Winter, Tobias Mann, Hans-Martin Haase, Leon Kuchenbecker and Katharina Jahn. Gecko3 is distributed under the LGPL, Version 3.0. Logo designed by Franziska Hufsky.