Im kommenden Semester wird statt Sequenzanalyse das Modul Algorithmische Phylogenetik angeboten. Sequenzanalyse wird dann wieder im SoSe 2026 zu hören sein.
News
SIRIUS 6.1.0 is Here!
We’re excited to announce the latest version of SIRIUS designed to improve your small molecule analysis workflow with a refreshed interface, streamlined processes, and several important fixes that ensure smoother performance and better data handling.
Highlights:
? New Color Scheme: A consistent and intuitive look throughout the entire identification process.
⚡ Streamlined Workflows: Tools are now automatically activated/deactivated to comply with SIRIUS workflow principles. You can also easily save and reload computation settings with our new preset function.
? Welcome Page Redesign: Get a quick overview of your account, connection details, and helpful resources to make the most of SIRIUS.
? Improved Result Views: Numerous enhancements and fixes across result views for a smoother analysis experience.
Times are changing but order matters
Our preprint Times are changing but order matters: Transferable prediction of small molecule liquid chromatography retention times is finally available on chemRxiv! Congratulations to Fleming and all authors.
In short, we show that prediction of retention times is a somewhat ill-posed problem, as retention times vary substantially even for nominally identical condition. Next, we show that retention order is much better preserved; but even retention order changes when the chromatographic conditions (column, mobile phase, temperature) vary. Third, we show that we can predict a retention order index (ROI) based on the compound structure and the chromatographic conditions. And finally, we show that one can easily map ROIs to retention times, even for target datasets that the machine learning model has never seen during training. Even for chromatographic conditions (column, pH) that are not present in the training data. Even for “novel compounds” that were not in the training data. Even if all of those restrictions hit us simultaneously.
In principle, this means that transferable retention time prediction (for novel compounds and novel chromatographic conditions) is “solved” for reversed-phase chromatography. There definitely is room for improvement, but that mainly requires more training data, not better methods. For that, we need your help: We need you, the experimentalists, to provide your reference LC-MS datasets. It does not matter what compounds are in there, it does not matter what chromatographic setup you used; as long as it is references data measured from standards (meaning that you know exactly each compound in the dataset), and as long as you tell us the minimum metadata, this will improve prediction quality.
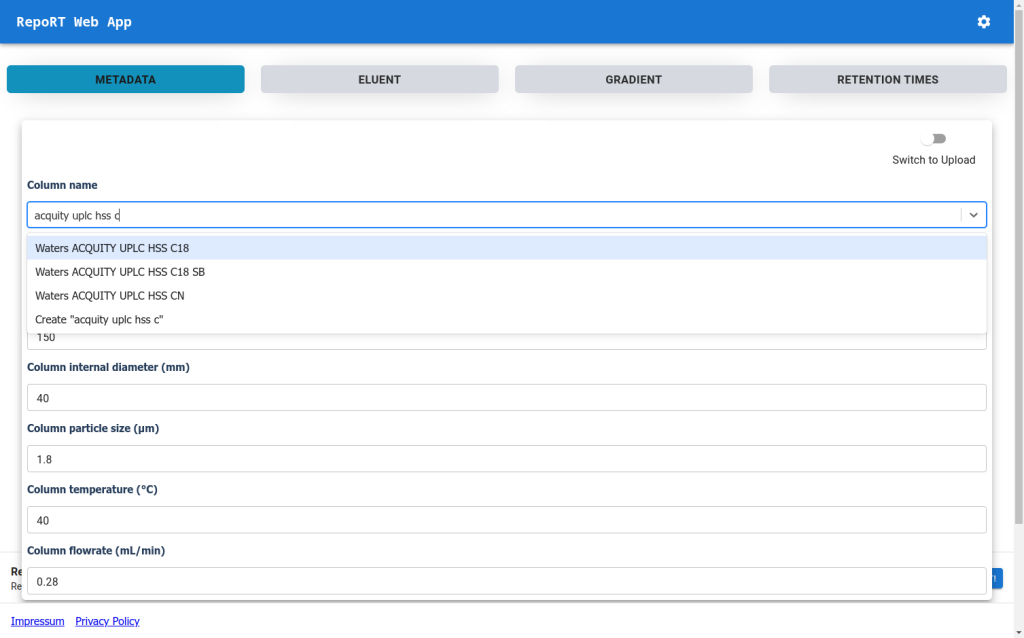
You can post your data wherever you like, but you can also upload them to RepoRT so that in the future, scientists from machine learning can easily access them. We now provide a web app that makes uploading really simple. If you are truly interested in retention time prediction, then this should be well-invested 30 min.
Meet Jonas at the IMPRS course on mass spectrometry in the MPI for Chemical Ecology
Jonas will give a lecture on SIRIUS at the IMPRS course “Introduction to mass spectrometry techniques and analysis” (2-4 December) in the Max Planck Institute for Chemical Ecology.
Meet us at GCB 2024 in Bielefeld
At GCB 2024 (30th Sep. – 2nd Oct.), Fleming will give a presentation about RepoRT. Jonas, Leopold and Roberto will show their posters in the poster session.
Meet Sebastian at the SETAC Asia-Pacific Biennial Meeting in Tianjin
Sebastian will give a plenary talk at the SETAC Asia-Pacific 14th Biennial Meeting (September 21 to 25, 2024).
Meet Sebastian at the SMAP conference in Lille
Sebastian will give a plenary talk at the conference on Mass Spectrometry and Proteomic Analysis (SMAP), which will be held from September 16 to 19, 2024.
New Web-App for RepoRT
To make the submission process to RepoRT easier, we have launched a web app, available under https://rrt.boeckerlab.uni-jena.de. Submitters can upload their data without having to adhere to specific formatting requirements. Metadata and gradient information can be entered directly online. Part of the data validation happens during the uploading process, providing immediate feedback when something is wrong. Inside the web app, submitters can also keep track of all submissions and their current status. As we continue to work on improving the web app, your feedback is of course very welcome!
Meet Sebastian at the Okinawa Workshop for Computational Mass Spectrometry
On June 24, Sebastian will give a presentation on SIRIUS 6 at the Okinawa Workshop for Computational Mass Spectrometry.
Meet Jonas at the Computational Metabolomics Workshop in Aberdeen
At the Computational Mass Spectrometry Workshop (19-21 June 2024), Jonas will hold a presentation on SIRIUS 6.
Meet Sebastian, Kai and Fleming at the Metabolomics conference in Osaka
At the Metabolomics 2024 (16-20 June), Sebastian will give a keynote presentation. Kai and Fleming will hold a workshop on SIRIUS 6.
Meet Markus at the ASMS conference in Anaheim
Meet Markus at the ASMS conference 2024 in Anaheim between June 2-6!
Meet Andrés and Nils at the European School of Metabolomics in Granada
Meet us at the EUSM 2024 in Granada! Nils will give a hands-on session on SIRIUS.
Prague workshop will be streamed
Good news for those who want to attend the Prague workshop but were not admitted: The Prague workshop on computational mass spectrometry will be streamed, see here. You should get the necessary software installed beforehand if you want to join in.
IMPRS call for PhD student
The International Max Planck Research School at the Max Planck Institute for Chemical Ecology in Jena is looking for PhD students. One of the projects (Project 7) is from our group on “rethinking molecular networks”. Application deadline is April 19, 2024.
Mass spectrometry (MS) is the analytical platforms of choice for high-throughput screening of small molecules and untargeted metabolomics. Molecular networks were introduced in 2012 by the group of Pieter Dorrestein, and have found widespread application in untargeted metabolomics, natural products research and related areas. Molecular networking is basically a method of visualizing your data, based on the observation that similar tandem mass spectra (MS/MS) often correspond to compounds that are structurally similar. Constructing a molecular network allows us to propagate annotations through the network, and to annotate compounds for which no reference MS/MS data are available. Since its introduction, the computational method has received a few “updates”, including Feature-Based Molecular Networks and Ion Identity Molecular Networks. Yet, the fundamental idea of using the modified cosine to compare tandem mass spectra, has basically remained unchanged at the core of the method.
In this project, we want to “rethink molecular networks”, replacing the modified cosine by other measures of similarity, including fragmentation tree similarity, the Tanimoto similarity of the predicted fingerprints, and False Discovery Rate estimates. See the project description for details.
We are searching for a qualified and motivated candidate from bioinformatics, machine learning, cheminformatics and/or computer science who want to work in this exciting, quickly evolving interdisciplinary field. Please contact Sebastian Böcker in case of questions. Payment is 0.65 positions TV-L E13.
IMPRS: https://www.ice.mpg.de/129170/imprs
MPI-CE: https://www.ice.mpg.de/
SIRIUS & CSI:FingerID: https://bio.informatik.uni-jena.de/software/sirius/
Literature: https://bio.informatik.uni-jena.de/publications/ and https://bio.informatik.uni-jena.de/textbook-algoms/
Jena is a beautiful city and wine is grown in the region:
https://www.youtube.com/watch?v=DQPafhqkabc
https://www.google.com/search?q=jena&tbm=isch
https://www.study-in.de/en/discover-germany/german-cities/jena_26976.php
Prague workshop on computational MS overbooked
Unfortunately, the Prague Workshop on Computational Mass Spectrometry (April 15-17, 2024) is heavily overbooked. The organizers will try to stream the workshop and the recorded sessions will be made available online, so check there regularly.
The workshop is organized by Tomáš Pluskal and Robin Schmid (IOCB Prague). Marcus Ludwig (Bright Giant) and Sebastian will give a tutorial on SIRIUS, CANOPUS etc.
Topics of the workshop are: MZmine, SIRIUS, matchms, MS2Query, LOTUS, GNPS, MassQL, MASST, and Cytoscape.
Meet Sebastian at the DGMS conference in Freising
Meet Sebastian at the conference of the Deutsche Gesellschaft für Massenspektrometrie (DGMS 2024) in Freising! The conference is March 10-13, and Sebastian will give a keynote talk on Tuesday, March 12.
BTW, another keynote will be given by our close collaboration partner Michael Witting, also on March 12.
RepoRT has appeared in Nature Methods
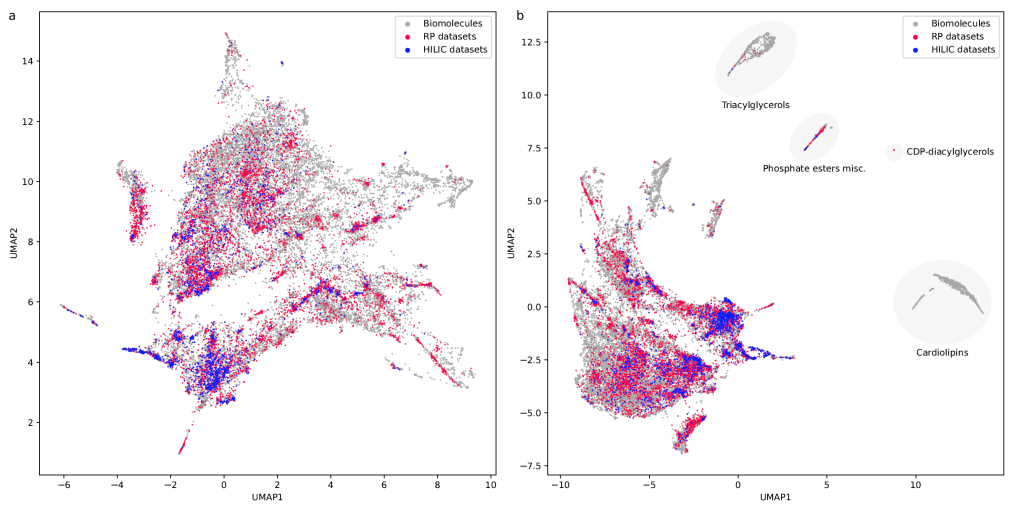
Our paper “RepoRT: a comprehensive repository for small molecule retention times” has just appeared in Nature Methods. This is joint work with Michael Witting (Helmholtz Zentrum München) as part of the DFG project “Transferable retention time prediction for Liquid Chromatography-Mass Spectrometry-based metabolomics“. Congrats to Fleming, Michael and all co-authors! In case you do not have access to the paper, you can find the preprint here and a read-only version here.
RepoRT is a repository for retention times, that can be used for any computational method development towards retention time prediction. RepoRT contains data from diverse reference compounds measured on different columns with different parameters and in different labs. At present, RepoRT contains 373 datasets, 8809 unique compounds, and 88,325 retention time entries measured on 49 different chromatographic columns using varying eluents, flow rates, and temperatures. Access RepoRT here.
If you have measured a dataset with retention times of reference compounds (that is, you know all the compounds identities) then please, contribute! You can either upload it to GitHub yourself, or you can contact us in case you need help. In the near future, a web interface will become available that will make uploading data easier. There are a lot of data in RepoRT already, but don’t let that fool you; to reach a transferable prediction of retention time and order (see below), this can only be the start.
If you want to use RepoRT for machine learning and retention time or order prediction: We have done our best to curate RepoRT: We have searched and appended missing data and metadata; we have standardized data formats; we provide metadata in a form that is accessible to machine learning; etc. For example, we provide real-valued parameters (Tanaka, HSM) to describe the different column models, in a way that allows machine learning to transfer between different columns. Yet, be careful, as not all data are available for all compounds or datasets. For example, it is not possible to provide Tanaka parameters for all columns; please see the preprint on how you can work your way around this issue. Similarly, not all compounds that should have an isomeric SMILES, do have an isomeric SMILES; see again the preprint. If you observe any issues, please let us know. See this interesting blog post and this paper as well as our own preprint on why providing “clean data” as well as “good coverage” are so important issues for small molecule machine learning.
Bioinformatische Methoden in der Genomforschung muss leider ausfallen
Nach aktuellem Kenntnisstand muss das Modul “Bioinformatische Methoden in der Genomforschung” im WS 23/24 leider ausfallen. Wir dürfen die Mitarbeiterstelle nicht besetzen, die wir dafür zwingend brauchen. Wir haben gekämpft und argumentiert und alles getan was wir konnten, aber am Ende war es leider vergeblich. Das Modul findet voraussichtlich das nächste Mal im WS 25/26 statt.
Warnung: Im Zuge der Sparmaßnamen an der FSU Jena kann es in Zukunft häufiger zu sollen kurzfristigen Ausfällen kommen.
Meet Sebastian at the Munich Metabolomics Meeting
Sebastian will give a tutorial on using SIRIUS and beyond at the Munich Metabolomics Meeting 2023.