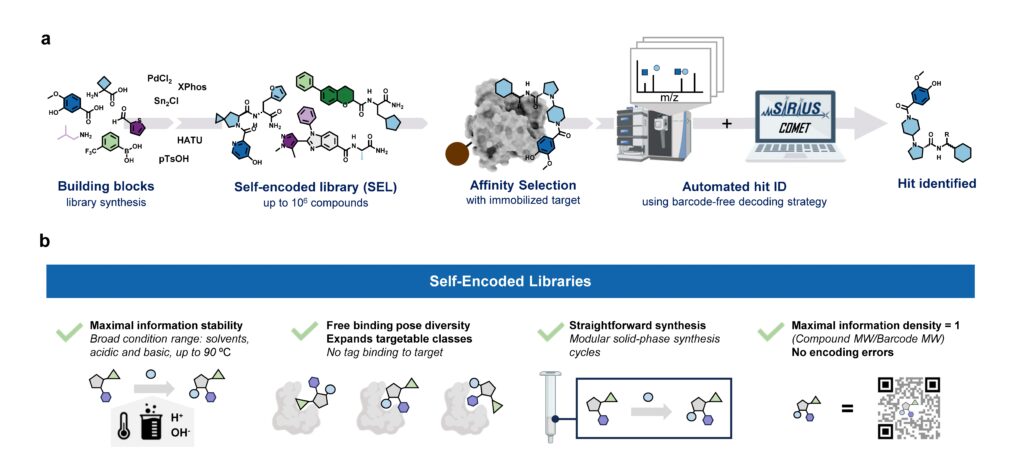
Our paper “Barcode-free hit discovery from massive libraries enabled by automated small molecule structure annotation” has finally been published in Nature Communications. This first publication from our joint project with Sebastian Pomplun (Leiden University and Oncode Institute) presents a barcode-free self-encoded library (SEL) platform that enables the screening of over half a million small molecules in a single experiment. Kudos to Sebastian Pomplun and his PhD student Edith van der Nol, and of course to all co-authors!
In general, discovering new pharmacologically active molecules that bind to a certain drug target is a tedious process as it requires the screening of very large compound libraries. In contrast to traditional high-throughput methods, affinity selection (AS) represents a powerful approach that allows the rapid screening of millions of molecules in just a single experiment. Here, the compound library is incubated with the receptor of interest, and in a following step, binders are separated from non-binders. This results in a final sample containing ideally only the binders which are called hits.
At this stage of the workflow, we know that this sample represents a subset of our initial screening library, but we do not know which molecules are actually in there. Therefore, the challenge remains in the identification of these hit compounds. This is most commonly done by initially attaching unique DNA barcodes to the molecules of the screening library, which can then be sequenced to identify the isolated hits. However, one of the fundamental drawbacks of these DNA-encoded libraries (DELs) is the potential interference of the attached tag during the affinity selection. This limitation becomes particularly problematic when the target protein has nucleic acid binding sites, making the screening against targets like transcription factors extremely difficult.
Consequently, an approach that does not require any tag while still allowing the simultaneous screening of a vast number of compounds is highly desirable. That is why our screening platform is based on affinity selection-mass spectrometry (AS-MS), which relies solely on MS to identify the selected hit compounds, eliminating the need for any tags. Thus, the name self-encoded libraries (SELs). Since our platform also features the use of split-&-pool libraries, containing potentially millions or even billions of combinatorially synthesized molecules, the monoisotopic mass alone is insufficient for the identification, and tandem mass spectrometry (MS/MS) has to be used instead. Therefore, we present a computational workflow, called COMET, for analyzing the acquired LC-MS/MS data and enabling the identification of the isolated hit compounds. Although the final AS sample contains only up to a few hundreds compounds, we observed that the corresponding LC-MS/MS data mainly contained features resulting from background noise, which usually lead to a large number of false-positive annotated compounds. We therefore implemented a filter into COMET that removes all features that do not match a genuine library molecule, either due to a non-matching precursor mass or fragmentation pattern.
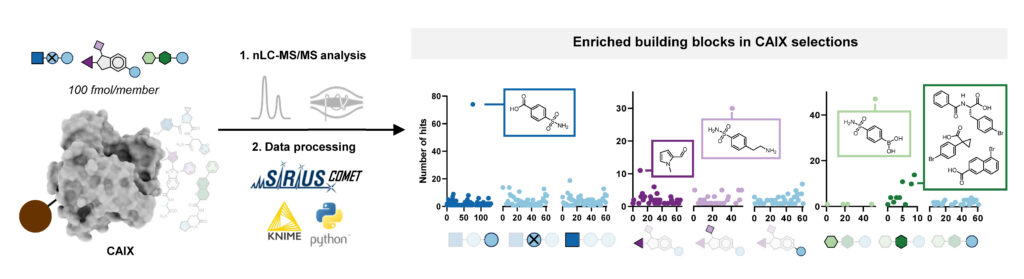
With this workflow in hand, Edith synthesized three combinatorial molecule libraries containing up to 750,000 compounds and incubated them separately with carbonic anhydrase IX (CAIX), a target typically used to benchmark novel DNA-encoded libraries. All three AS-MS experiments resulted in the identification of hit compounds containing a sulfonamide building block, which aligns well with the literature about known CAIX binders. Subsequent validation confirmed these hits’ binding affinity to CAIX, demonstrating the effectiveness of our screening platform.
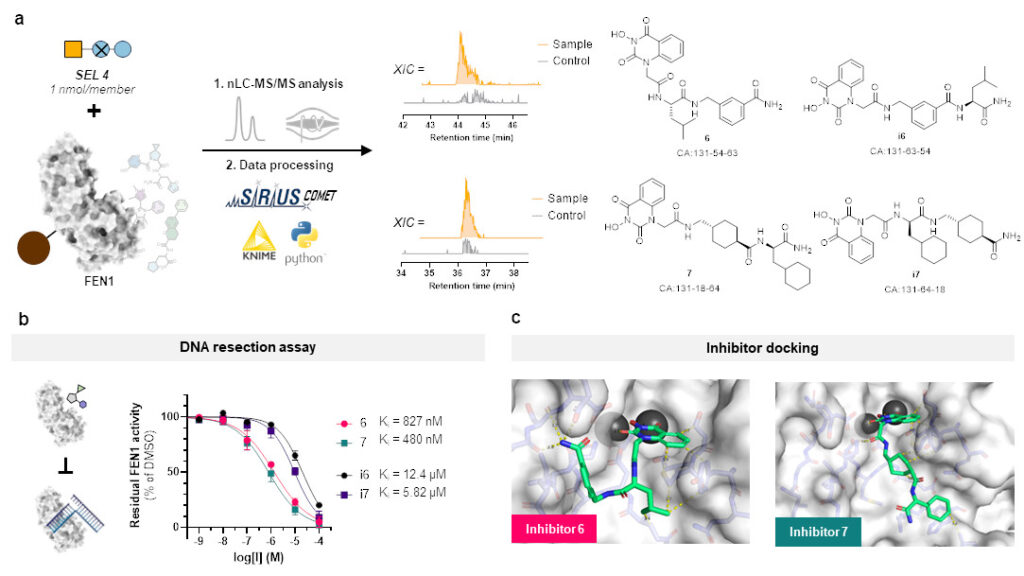
But this represents more or less only a sanity check and does not clearly show the advantage of our SEL platform compared to DELs. So, we chose a target protein which is beyond the scope of DELs: flap endonuclease 1 (FEN1), a DNA-processing enzyme essential for replication and repair, and over-expressed in multiple cancer types. For this challenging target, we synthesized a new, more focused library comprising 4,000 members after determining that our previous libraries were insufficient for clear hit identification. This approach successfully yielded four hits, all of which demonstrated inhibitory activity against FEN1.
All in all, our SEL-COMET workflow represents a rapid, barcode-free screening approach for early drug discovery, and we hope that this approach will be widely adopted in both academic and industrial research settings.