The discovery of pharmacologically active molecules that effectively target specific drug receptors necessitates the screening of millions of compounds. In contrast to traditional high-throughput methods, affinity selection-based strategies enable the rapid screening of entire compound libraries in a single experiment. This technique involves incubating a compound library with the drug target of interest, followed by a physical separation step to separate the molecules that have bound to the target from those that remain unbound. However, the main challenge remains in the identification of these isolated hit compounds [1].
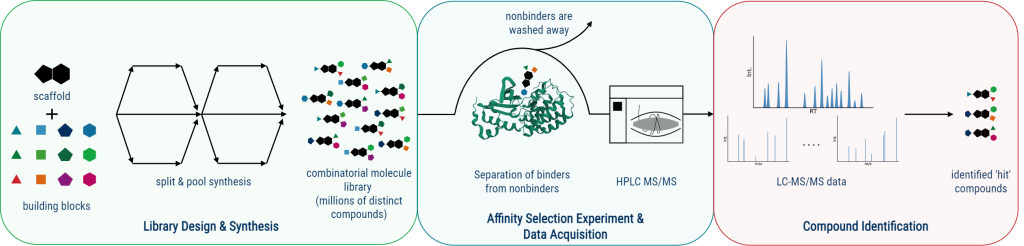
In this project, our focus centers on affinity selection-mass spectrometry (AS-MS) which relies on mass spectrometry to facilitate the identification of these compounds. Since we consider combinatorial molecule libraries, often comprising millions or even billions of distinct molecules, we are compelled to use tandem mass spectrometry (MS/MS), as the monoisotopic mass of the molecules is likely to be insufficient for their identification. Therefore, the overarching goal of this project is the development of a computational method that allows the identification of these isolated hit compounds from the acquired MS/MS data.
Acknowledgements
We gratefully thank the Thüringer Aufbaubank, which is financing this project on behalf of the Free State of Thuringia and with funds from the European Union as part of the European Regional Development Fund (ERDF Program 2021-2027 Thuringia).
References:
- J. M. Mata et al. Advances in Ultrahigh Throughput Hit Discovery with Tandem Mass Spectrometry Encoded Libraries. J. Am. Chem. Soc., 2023
