COSMIC is a workflow that allows you to assign confidence to structure annotations. For every structure annotated by CSI:FingerID, COSMIC provides a confidence score (a number between 0 and 1) that tells you how likely it is that this annotation is correct. This is similar in spirit to what is done in spectral library search: Not only is the cosine score used to decide which candidate best fits to the query spectrum; in addition, we use the cosine of the top-scoring candidate (the hit) to decide whether it is likely correct (say, above 0.8), incorrect (say, below 0.6) or in the “twilight” in-between. If you have been using CSI:FingerID for some time, you might have noticed that finding such thresholds is not possible for the CSI:FingerID score. COSMIC closes this gap and tells you if an annotation is likely correct or incorrect. COSMIC will soon be integrated in SIRIUS.
Installation
COSMIC confidence scores have been integrated into SIRIUS 4.8 and above. Download the appropriate SIRIUS version for your operating system here. The GUI version (which also includes the full command line version) requires no external JRE. Everything is included: Download, extract, execute. Average installation times should be a few minutes.
Usage
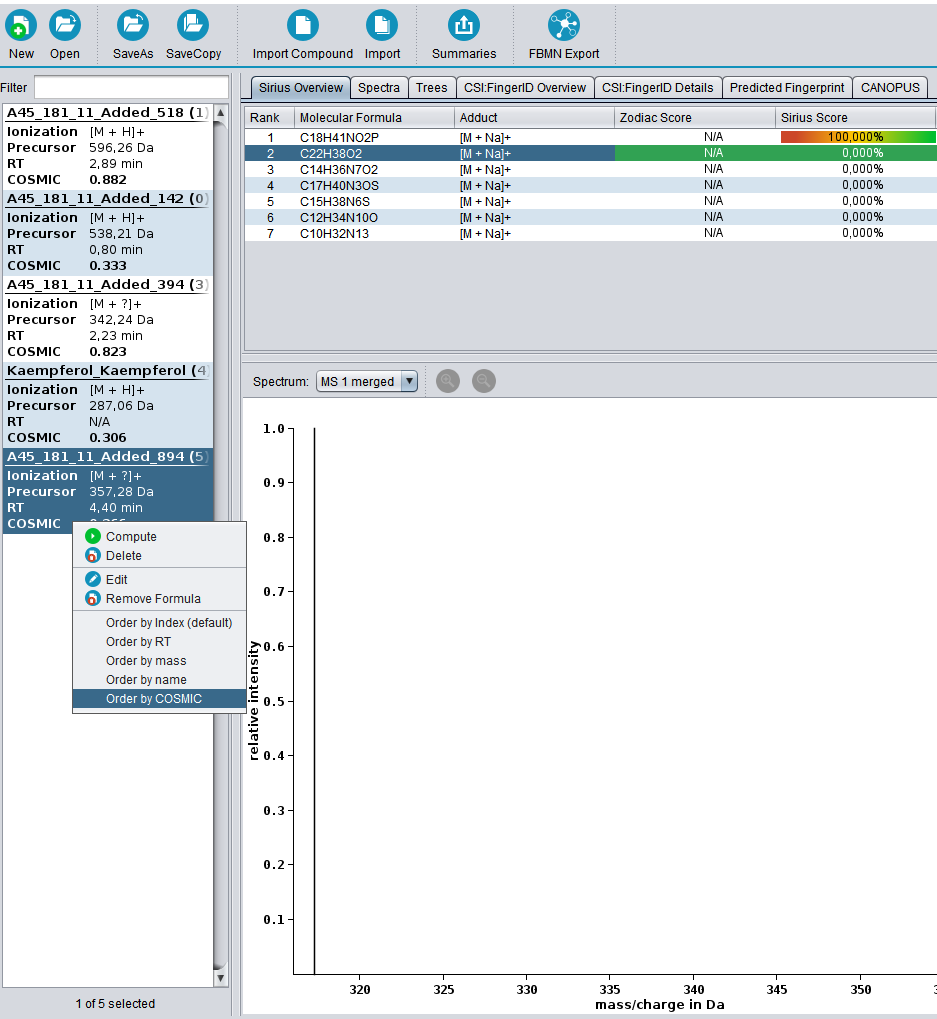
Calculating COSMIC confidence scores is parameter free and will be executed automatically every time a CSI:FingerID search is performed. COSMIC scores for a compound are shown in the compound list on the left. The COSMIC demo data contains five compounds with varying confidence values when performing SIRIUS + CSI:FingerID (biological database). Drag and drop the files into the GUI, and press “compute all” as shown. Expected run time for the small demo dataset should only be a few minutes.
The compound list can be sorted by COSMIC score by right clicking it and selecting “Order by COSMIC”. Detailed documentation on how to run SIRIUS + CSI:FingerID can be found in the SIRIUS documentation.
FAQ
How should i interpret COSMIC confidence values?
COSMIC confidence scores need to be interpreted with some care. It is important to understand, that they do not represent probabilities, and thus there is no statistical interpretation of a confidence value. If doing large scale analysis, one should focus on the highest confident hits (for example the Top 1/5/10%), (mostly) regardless of their confidence value.
An annotation received a low COSMIC score, even though am i sure it is correct!
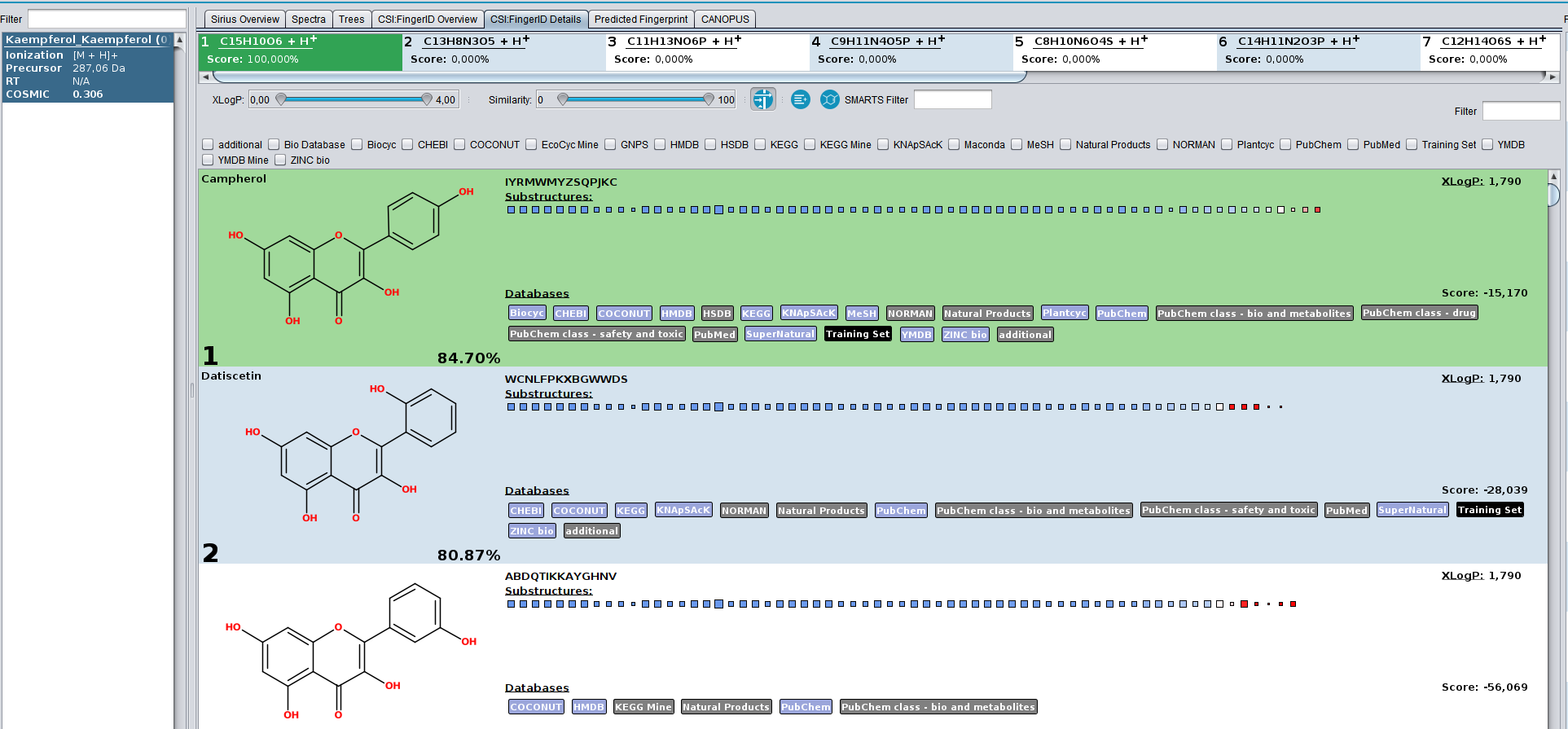
In this example, the spectrum of Campherol from the SIRIUS demo data receives the low COSMIC score of 0.3, even though the structure annotation is correct. The reason for this is, that the database we searched in contains multiple extremely similar structures, that have very similar fingerprint representations as well as CSI:FingerID scores. If the true structure was not know, each of these highly-similar compounds could be the correct structure. Hence, the hit receives a low confidence. This is not a limitation of CSI:FingerID and COSMIC but rather of mass spectrometry. The MS/MS of these structures will look very similar.
Use cases
Searching in hypothetical structure databases
Apart from enhancing your previous CSI:FingerID experience by telling you if an annotation is likely correct or not, COSMIC makes searching in hypothetical databases viable. We demonstrate this, by generating a database of hypothetical bile acid structures, combinatorially adding amino acids to bile acid cores, yielding 28,630 plausible bile acid conjugate structures. We then searched query MS/MS data from a mice fecal dataset in this combinatorial database, and used the COSMIC confidence score to distinguish between hits that are likely correct or incorrect. We manually evaluated the top 12 hits and found that 11 annotation (91.6%) were likely correct; two annotations were further confirmed using synthetic standards. All 11 bile acid conjugates are “truly novel”, meaning that we could not find those structures in PubChem or any other structure database (or publication). Whereas reporting 11 novel bile acid conjugates may appear rather cool, we argue it is even cooler that we did this without a biological hypothesis beyond “there might be bile acid conjugates out there which nobody knows about”; and that COSMIC found the top bile acid conjugate annotations in a fully automated manner.
Flipping the metabolomics workflow
Additionally, COSMIC enables you to “flip the workflow”: Annotate large quantities of data, then look at novel compound annotations with high confidence and form your hypothesis from there! To demonstrate this ability, we have also annotated 2,666 LC-MS/MS runs from human samples with molecular structures which are currently absent from HMDB, and for which no MS/MS reference data are available; and finally, 17,414 LC-MS/MS runs with annotations for which no MS/MS reference data are available (see the COSMIC preprint for details)
References
Data availability
- Scripts for generating the bile acid conjugate database
- Link to HUMAN and ORBITRAP dataset results
- Download link for the spectral library generated from the Bile acid conjugate study (structure annotations are putative)
- Download link for the spectral library generated from the human dataset study (structure annotations are putative)
- Download link for the spectral library generated from the orbi dataset study (structure annotations are putative)
- Biomolecule structure database (SMILES)