Our paper “RepoRT: a comprehensive repository for small molecule retention times” has just appeared in Nature Methods. This is joint work with Michael Witting (Helmholtz Zentrum München) as part of the DFG project “Transferable retention time prediction for Liquid Chromatography-Mass Spectrometry-based metabolomics“. Congrats to Fleming, Michael and all co-authors! In case you do not have access to the paper, you can find the preprint here and a read-only version here.
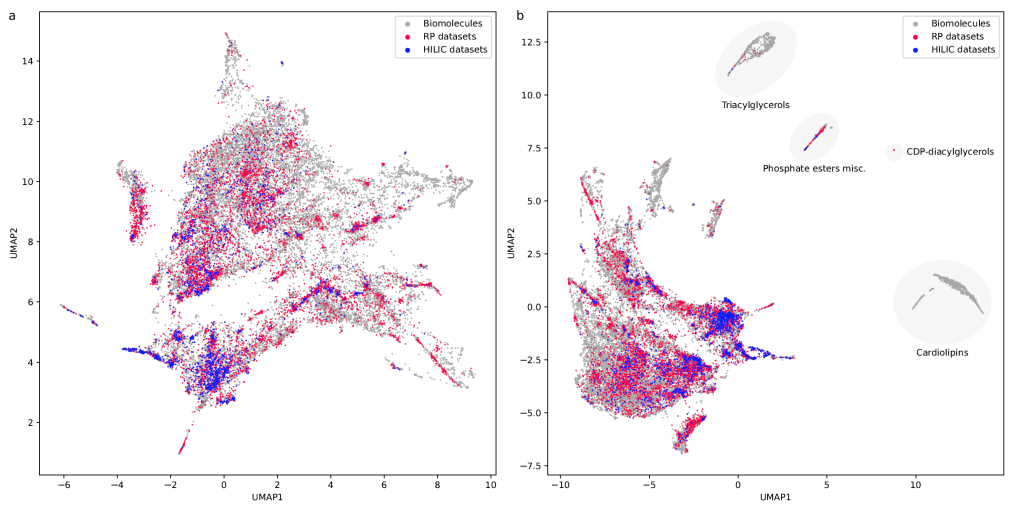
RepoRT is a repository for retention times, that can be used for any computational method development towards retention time prediction. RepoRT contains data from diverse reference compounds measured on different columns with different parameters and in different labs. At present, RepoRT contains 373 datasets, 8809 unique compounds, and 88,325 retention time entries measured on 49 different chromatographic columns using varying eluents, flow rates, and temperatures. Access RepoRT here.
If you have measured a dataset with retention times of reference compounds (that is, you know all the compounds identities) then please, contribute! You can either upload it to GitHub yourself, or you can contact us in case you need help. In the near future, a web interface will become available that will make uploading data easier. There are a lot of data in RepoRT already, but don’t let that fool you; to reach a transferable prediction of retention time and order (see below), this can only be the start.
If you want to use RepoRT for machine learning and retention time or order prediction: We have done our best to curate RepoRT: We have searched and appended missing data and metadata; we have standardized data formats; we provide metadata in a form that is accessible to machine learning; etc. For example, we provide real-valued parameters (Tanaka, HSM) to describe the different column models, in a way that allows machine learning to transfer between different columns. Yet, be careful, as not all data are available for all compounds or datasets. For example, it is not possible to provide Tanaka parameters for all columns; please see the preprint on how you can work your way around this issue. Similarly, not all compounds that should have an isomeric SMILES, do have an isomeric SMILES; see again the preprint. If you observe any issues, please let us know. See this interesting blog post and this paper as well as our own preprint on why providing “clean data” as well as “good coverage” are so important issues for small molecule machine learning.