We are happy to announce that a major version upgrade of SIRIUS is available! Scroll to the bottom to get a visual impression of the changes.
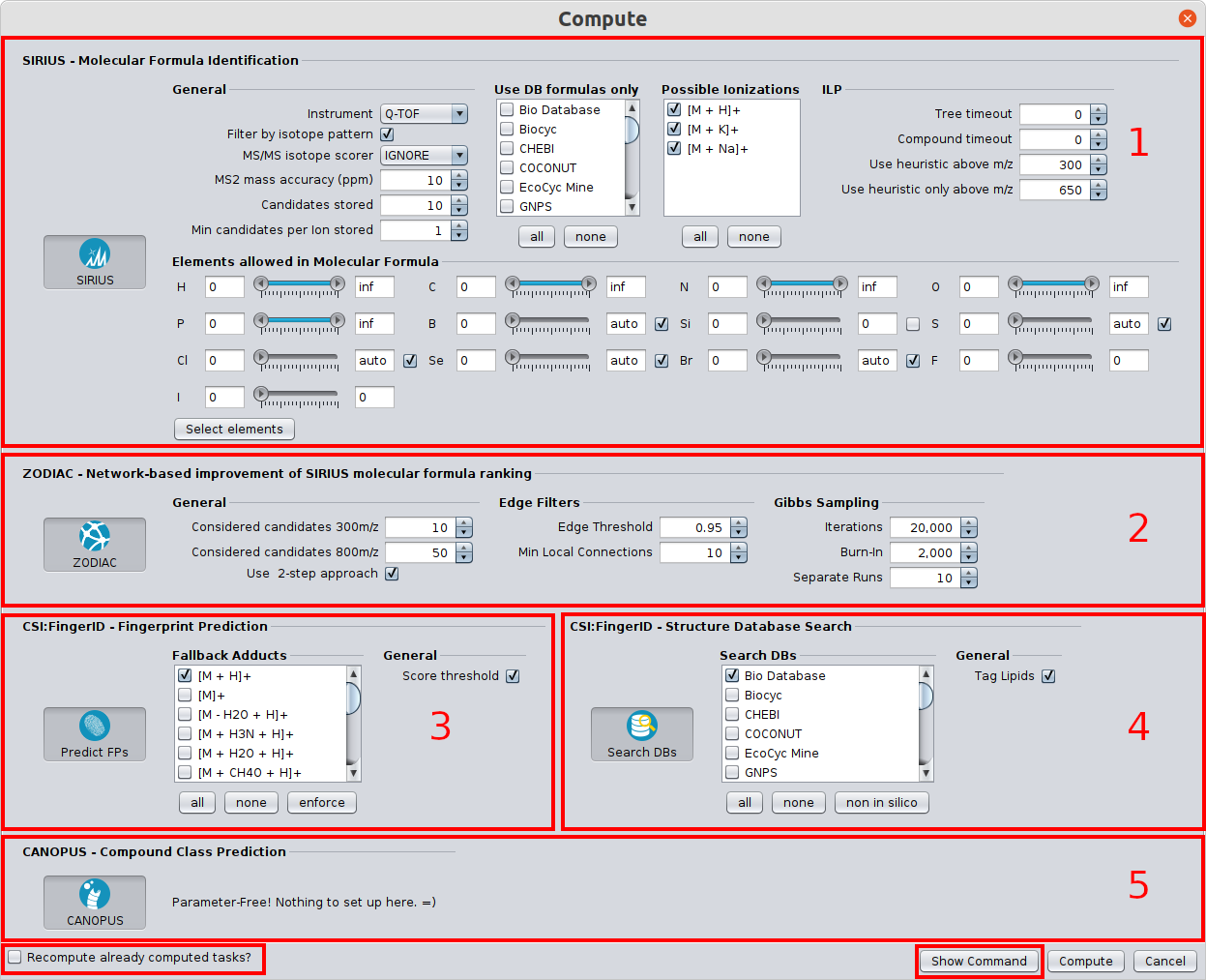
SIRIUS 5 now includes the following new features and improvements:
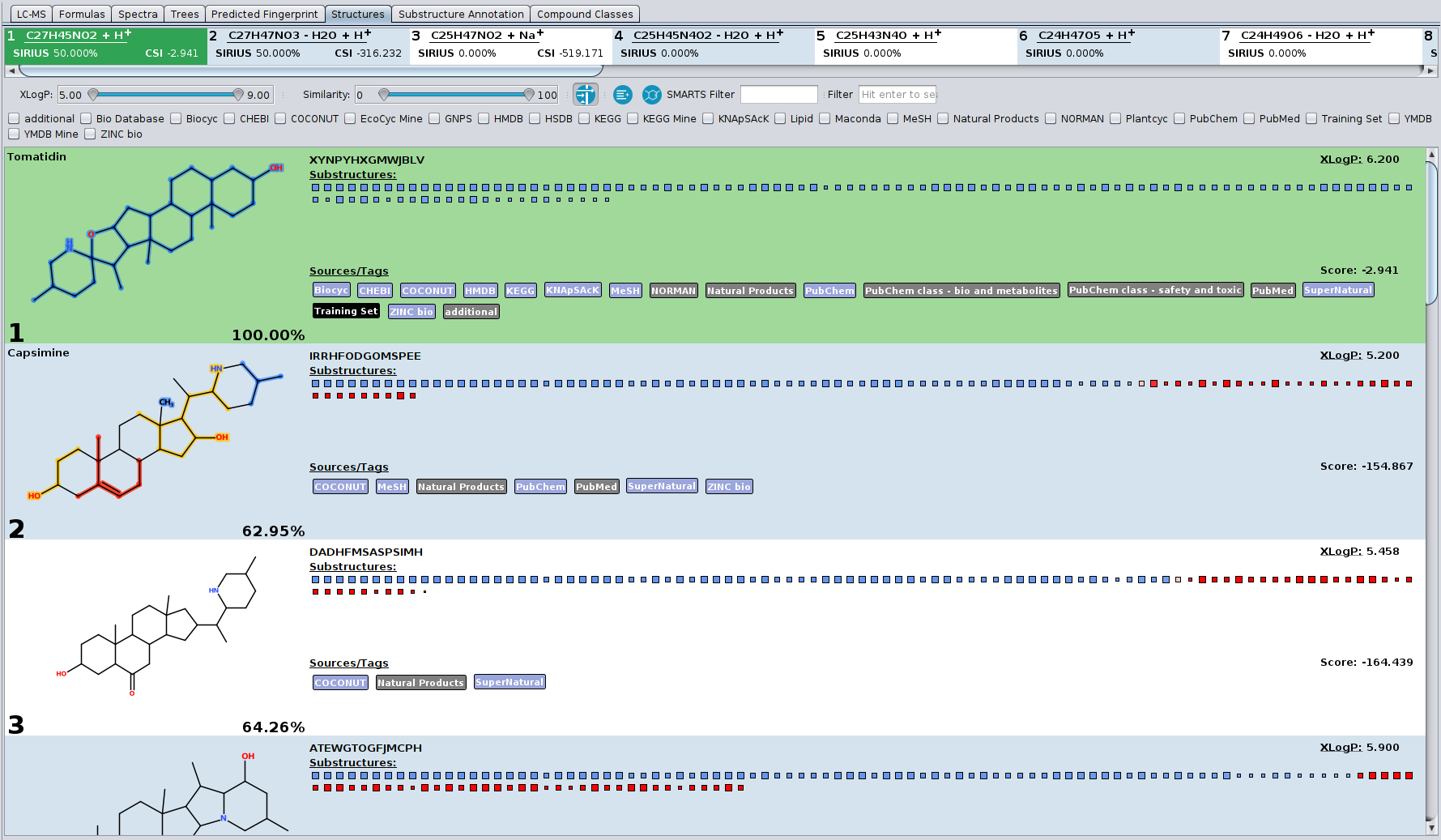
- Lipid class annotation with El Gordo: Lipid structures that have the same molecular formula (usually belonging to the same lipid class) can be extremely similar to each other, often only differing in the position of the double bonds. These extremely similar structures may be even not differentiable by mass spectrometry at all. SIRIUS 5 now predicts the lipid class from the spectrum; when performing CSI:FingerID database search, all structure candidates that belong to this lipid class are tagged.
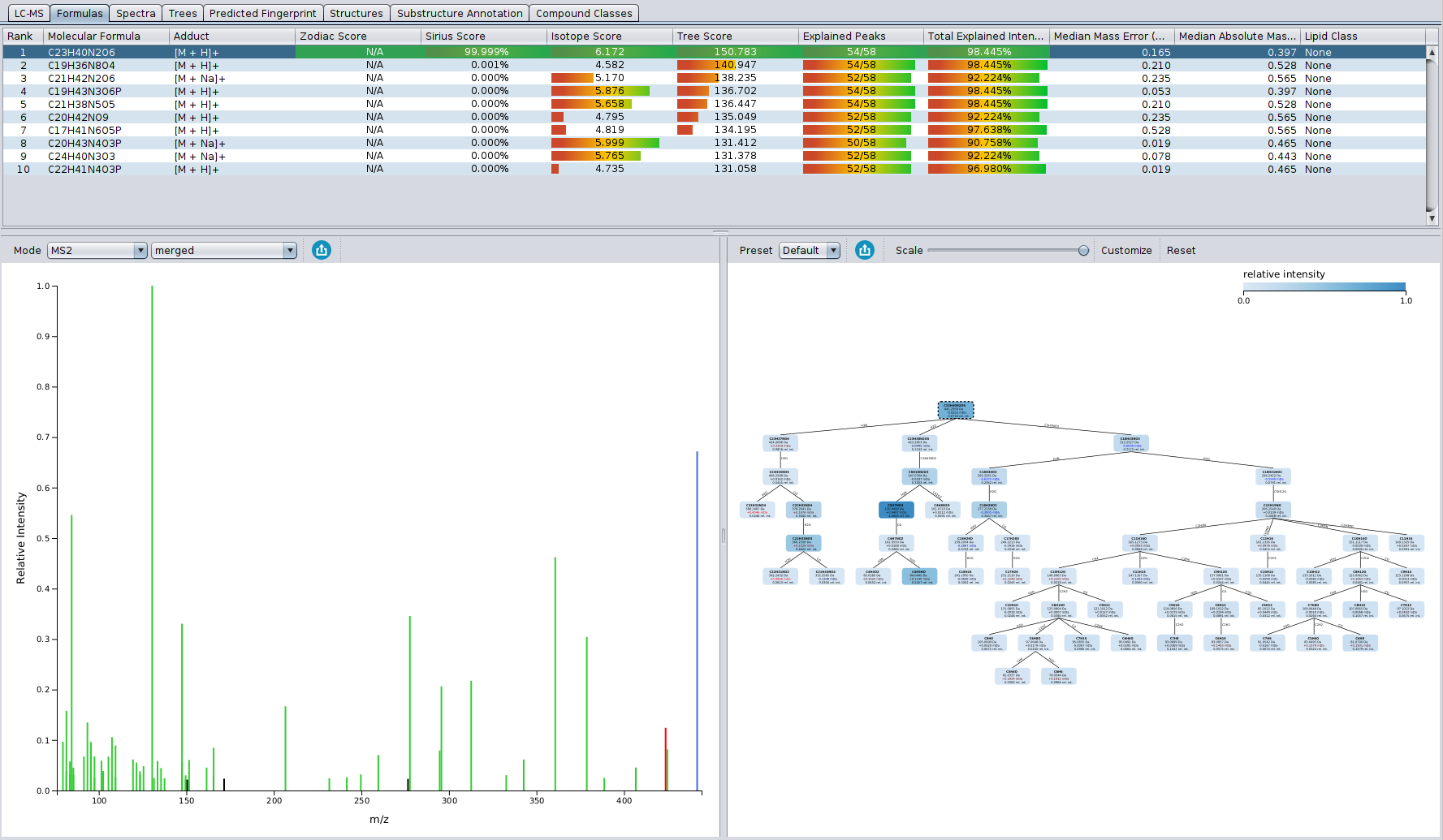
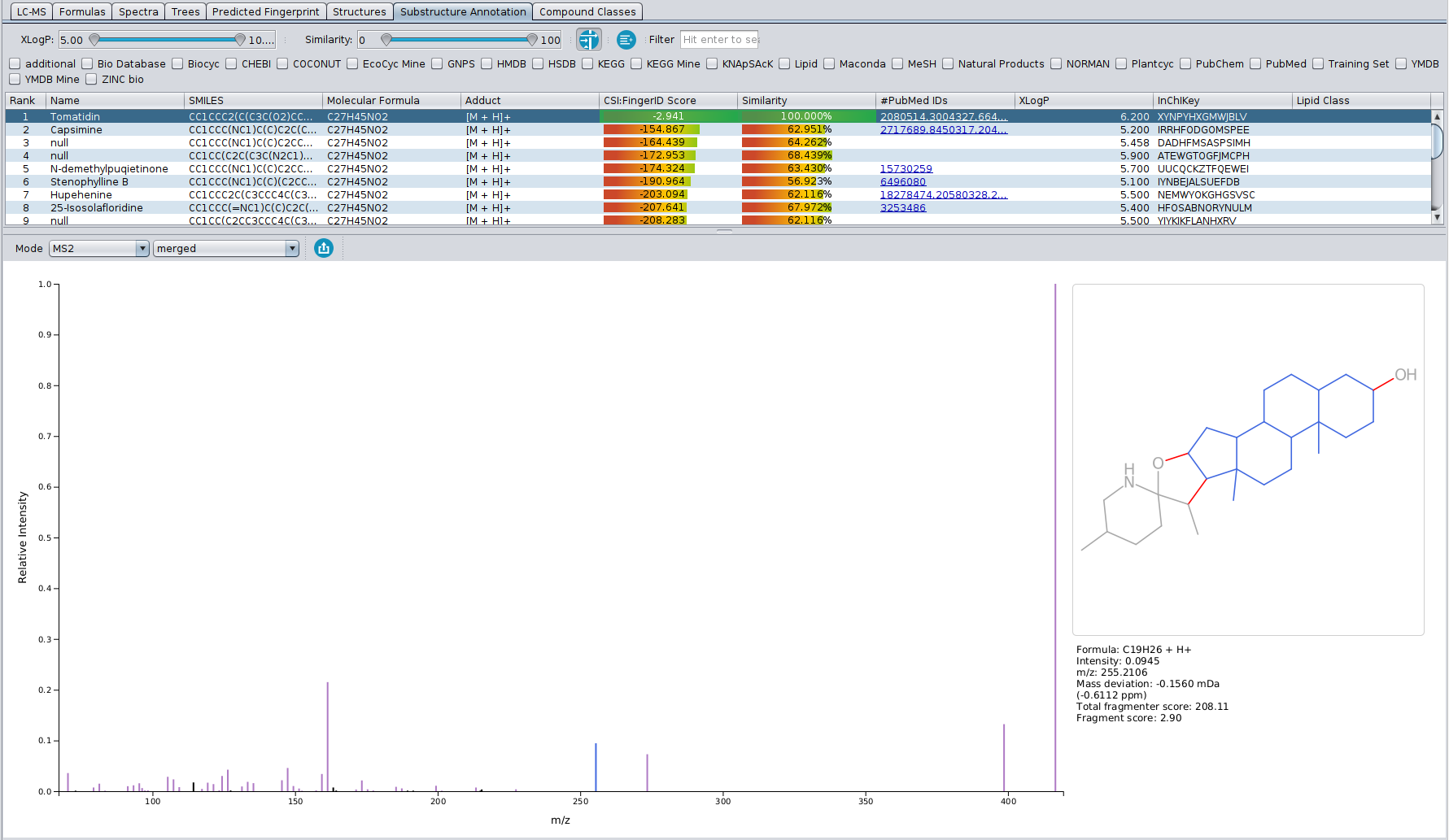
- Sub-structure annotation with Epimetheus: For experimentalists, CSI:FingerID search may seem like a black box. If you want to perform manual validation of CSI:FingerID structure candidates, Epimetheus now provides a direct connection between structure candidates and your input MS/MS spectra. Sub-structures of the structure candidate of your choice are generated by a combinatorial fragmenter and assigned to peaks in the MS/MS spectrum. The new Epimetheus view allows you to directly visualize and inspect these sub-structure annotations.
- Feature-rich spectrum viewer: Improved functionality of the SIRIUS spectrum viewer, including mirror plots of measured vs predicted isotope pattern.
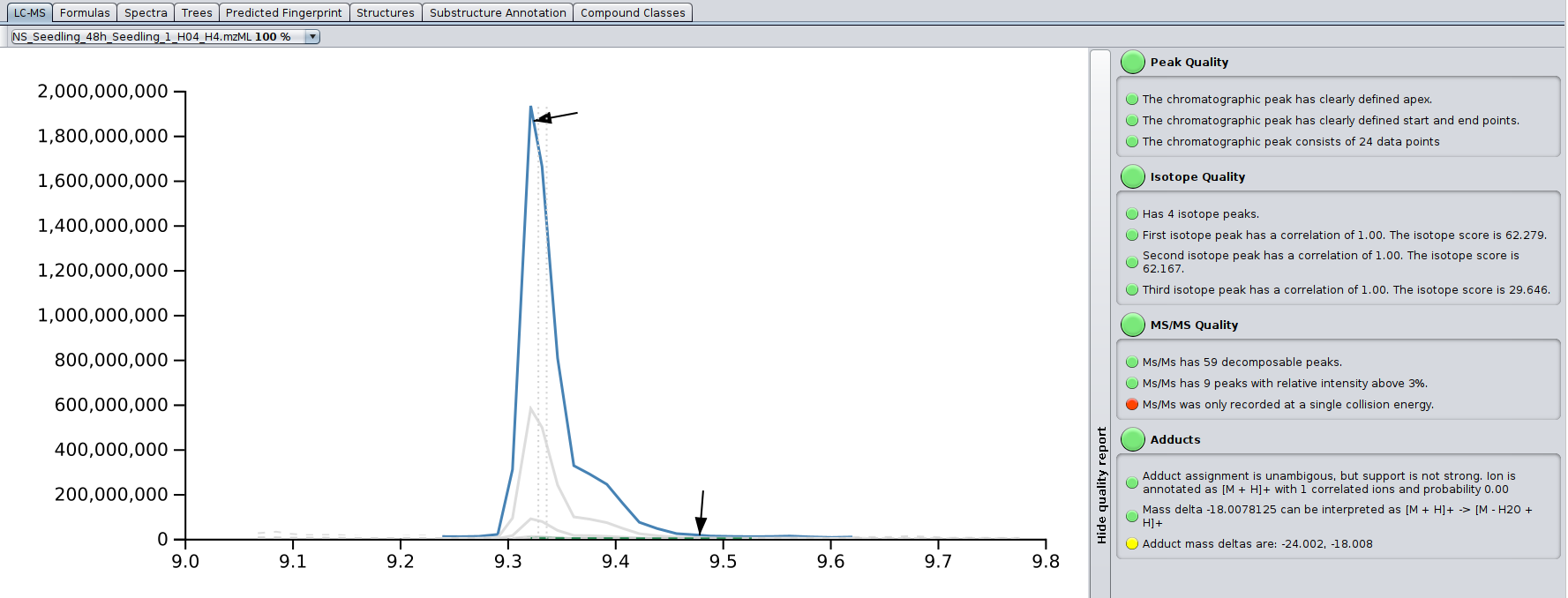
- New LC-MS view: A new view in SIRIUS 5 that shows the extracted-ion chromatogram of a compound, including its detected adducts and isotopes. A traffic light allows for quick spectral quality assessment.
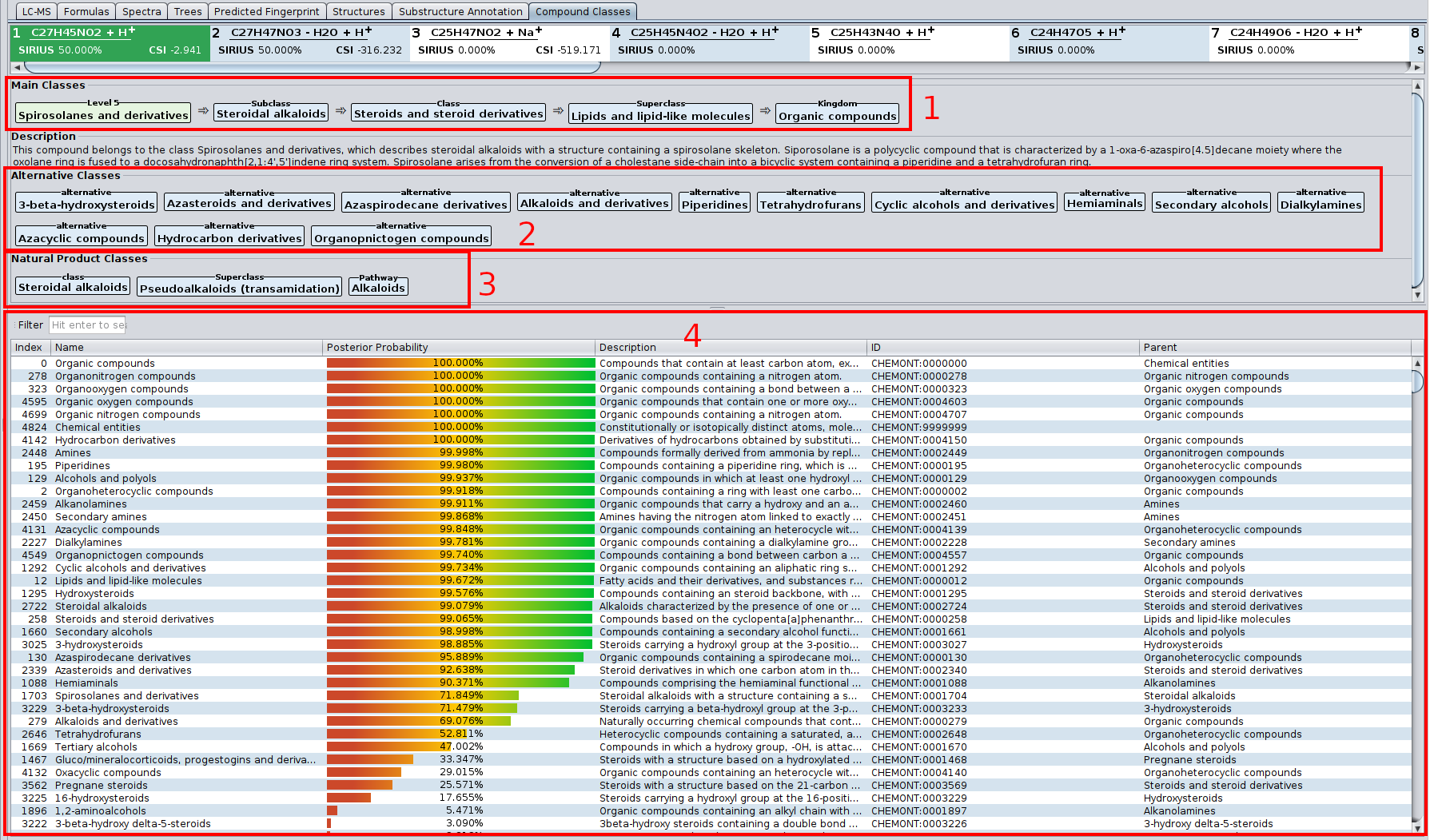
- CANOPUS now fully supports Natural Product Classes (NPC).
- Advanced filtering options on the compound list, including the new lipid class annotations.
- Support for additional spectrum file formats (.msp, massbank, .mat)
If you were previously using SIRIUS 4, please be aware of the following breaking changes:
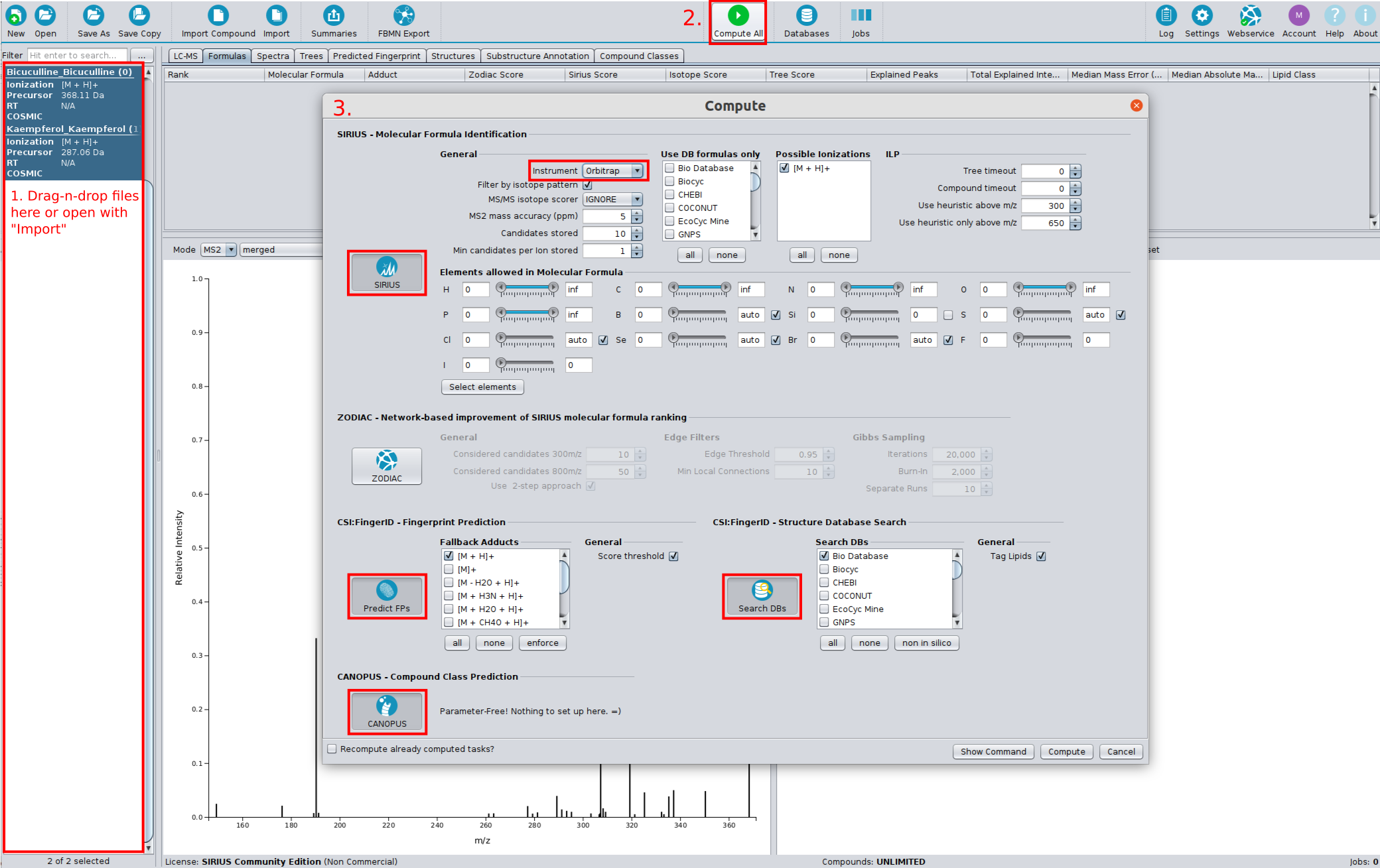
- User authentication: A user account and license is now needed to use the online features of SIRIUS. The license is free and automatically available for non-commercial use. If your account is not automatically verified because your non-commercial research institution is not whitelisted yet, please contact us at sirius@uni-jena.de.
- New project space compression: Method level directories are now compressed archives to reduce number of files and save storage.
- Changed summary writing: Summary writing has been made a separate sub-tool (
write-summaries). Summary files format has slightly changed. - Prediction / DB search split: The
fingerid/structuresub-tool has been split into afingerprint(fingerprint prediction) and astructure(structure db search) sub-tool. This allows the user to recompute the database search without having to recompute the fingerprint and compound class predictions. It further allows to compute CANOPUS compound class prediction without having to perform structure db search. - Updated fingerprints: Updated fingerprint vector. Fingerprint related results of SIRIUS 4 projects may have to be recomputed to perform certain analysis steps (e.g. recompute db-search). Reading the projects is still possible and formula results are not affected.
- Custom database format change: Custom database format has changed. Custom databases need to be re-imported.
- GUI column rename: Some views in the GUI have been renamed to better reflect their position and role in the workflow.
For a quick overview on these new features and changes, visit our YouTube channel. Please refer to our online documentation for a more comprehensive overview and help us squash all remaining bugs by contributing at our GitHub!