We’re excited to announce the latest version of SIRIUS designed to improve your small molecule analysis workflow with a refreshed interface, streamlined processes, and several important fixes that ensure smoother performance and better data handling.
Highlights:
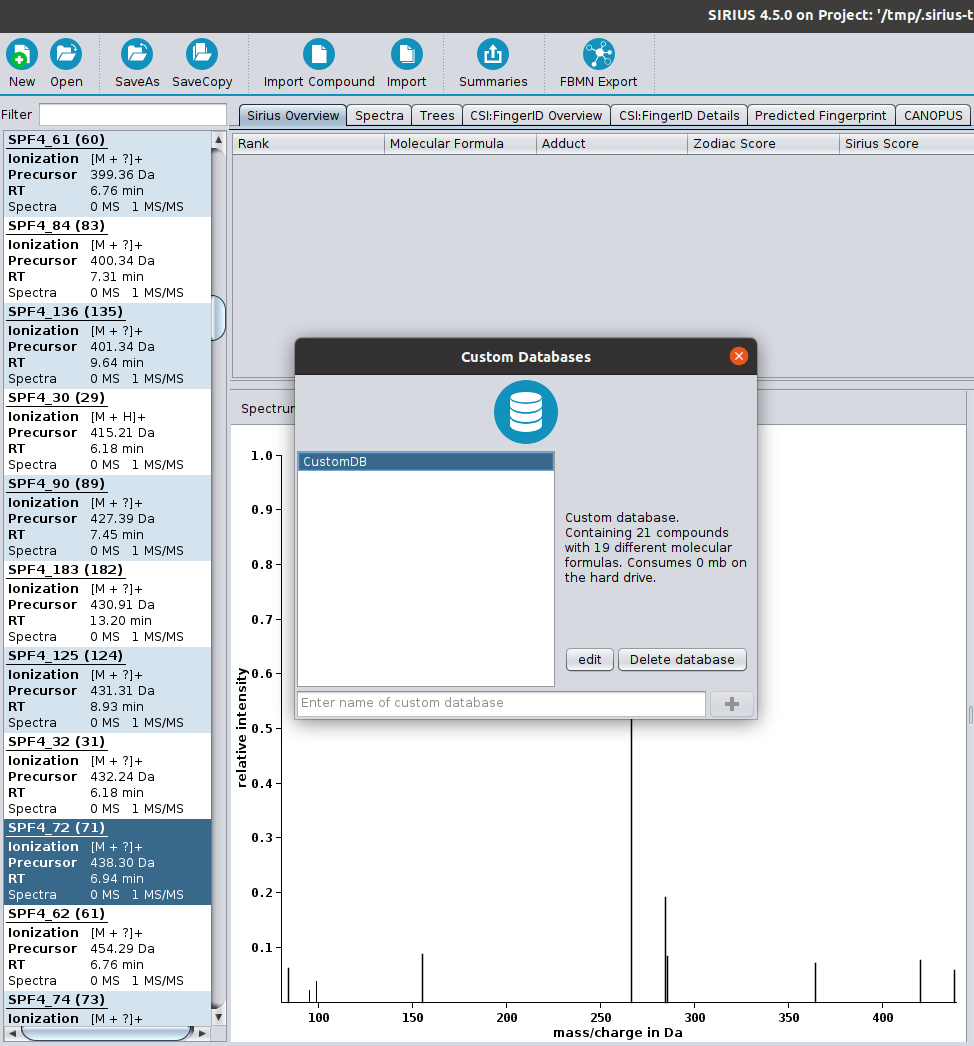
? New Color Scheme: A consistent and intuitive look throughout the entire identification process.
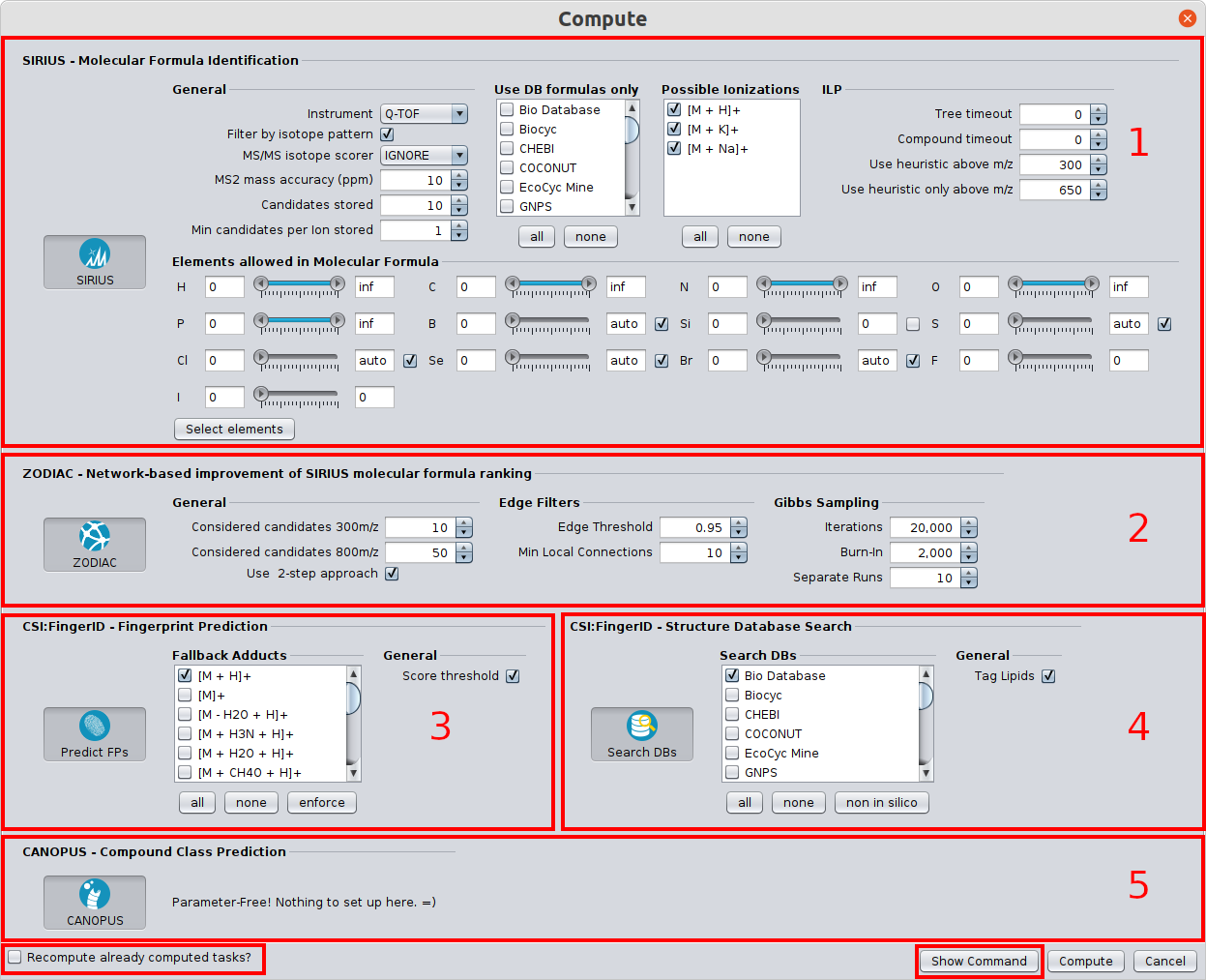
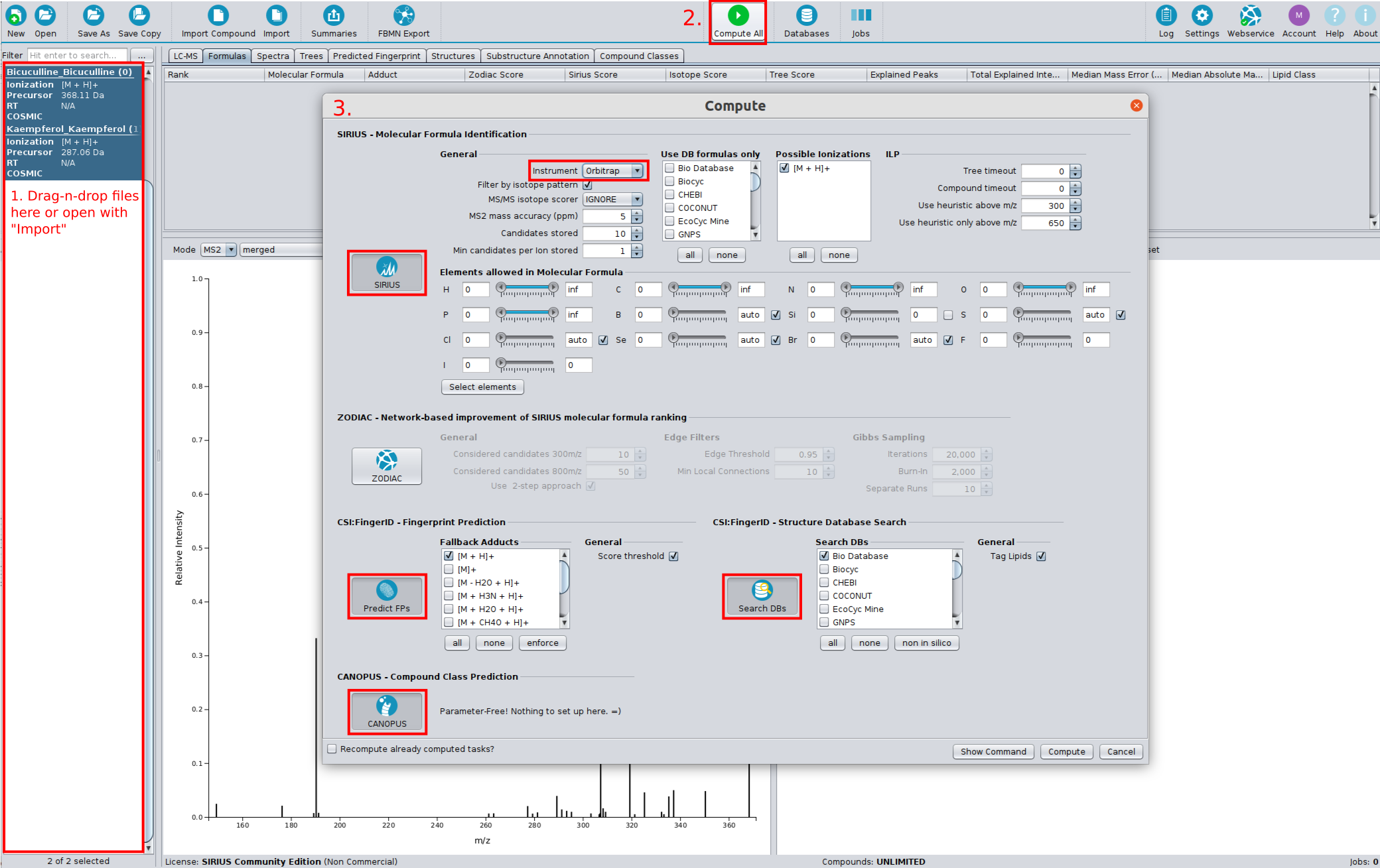
⚡ Streamlined Workflows: Tools are now automatically activated/deactivated to comply with SIRIUS workflow principles. You can also easily save and reload computation settings with our new preset function.
? Welcome Page Redesign: Get a quick overview of your account, connection details, and helpful resources to make the most of SIRIUS.
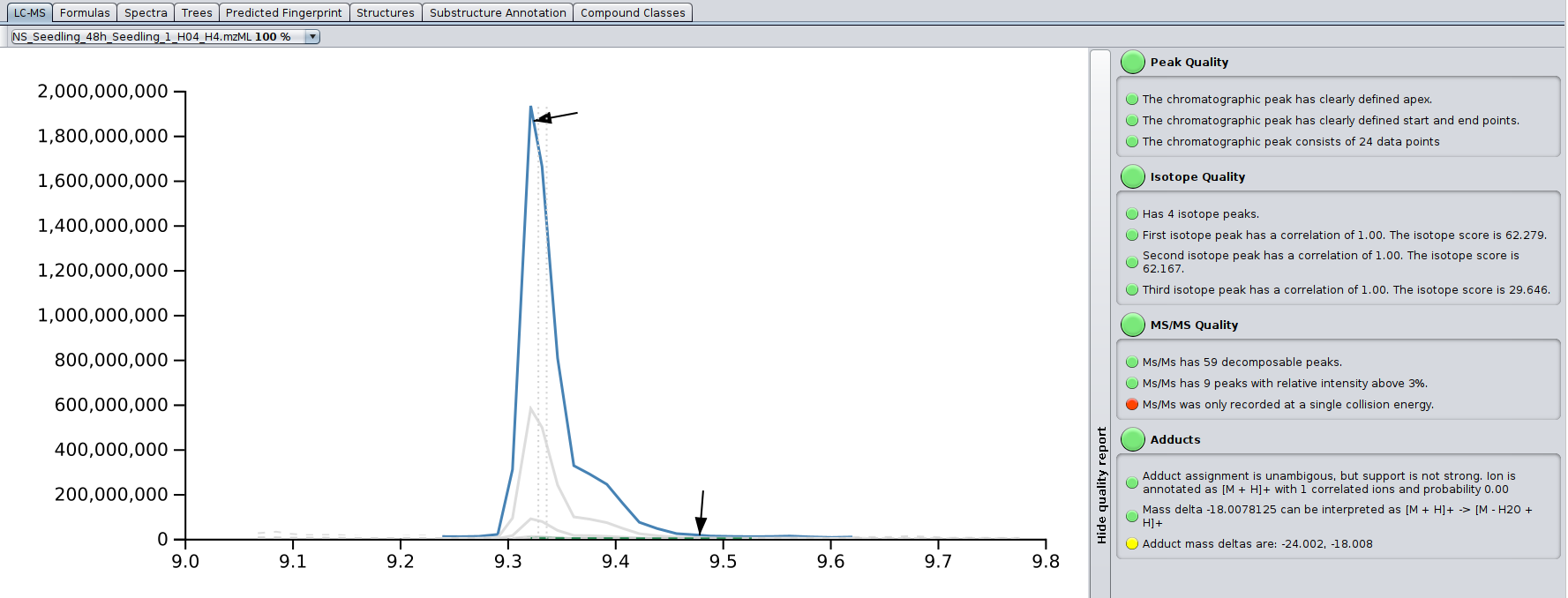
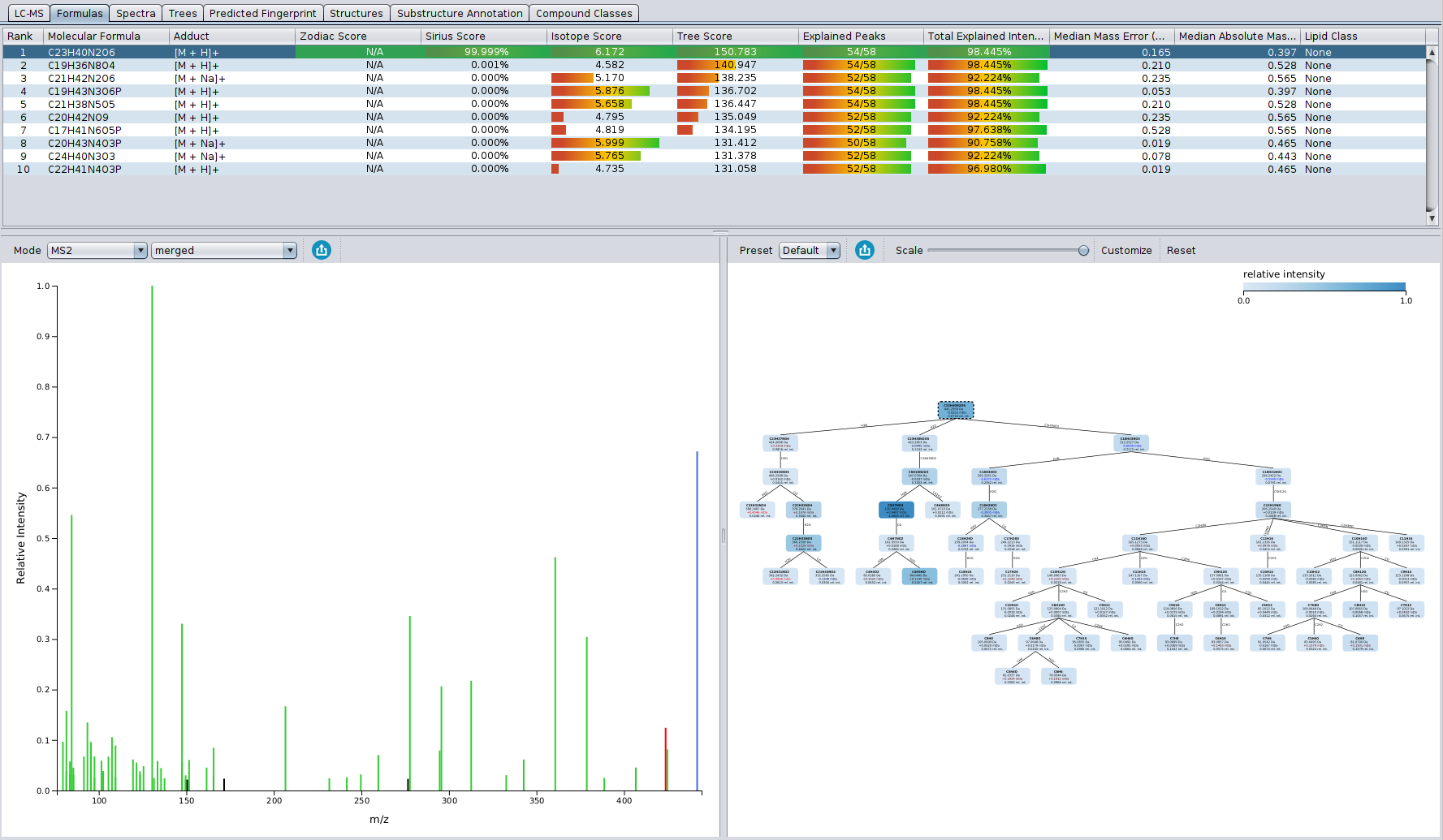
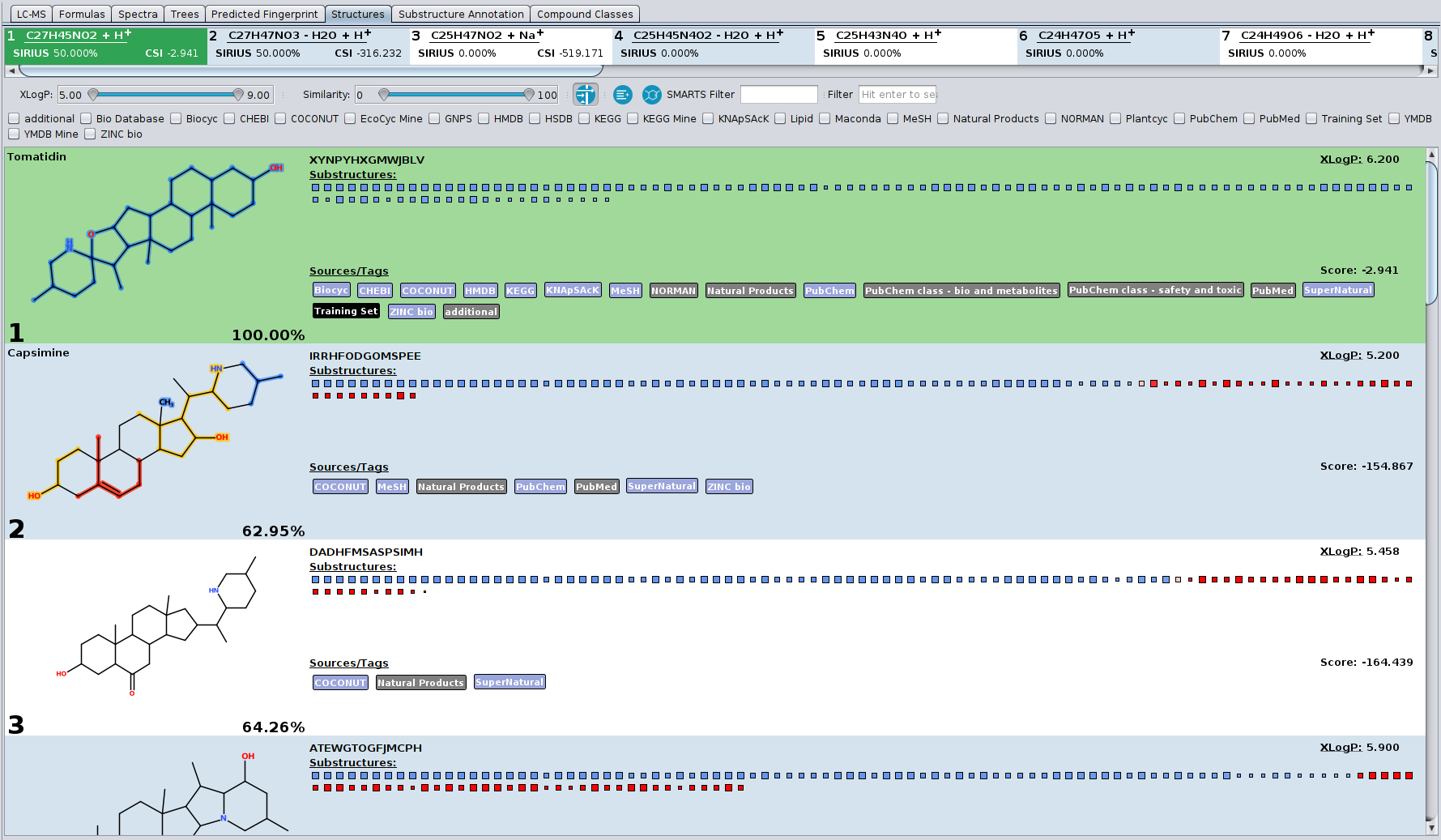
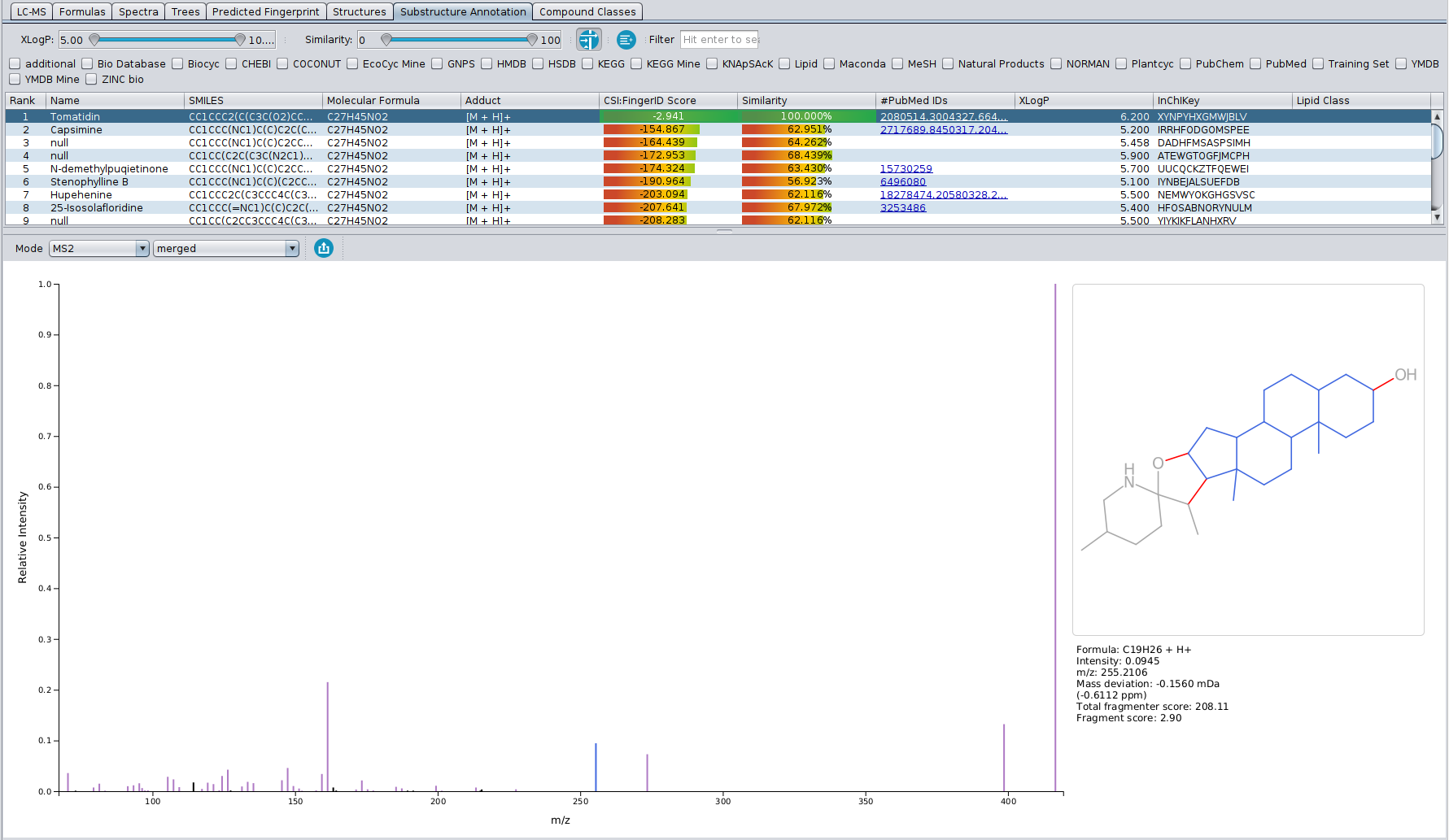
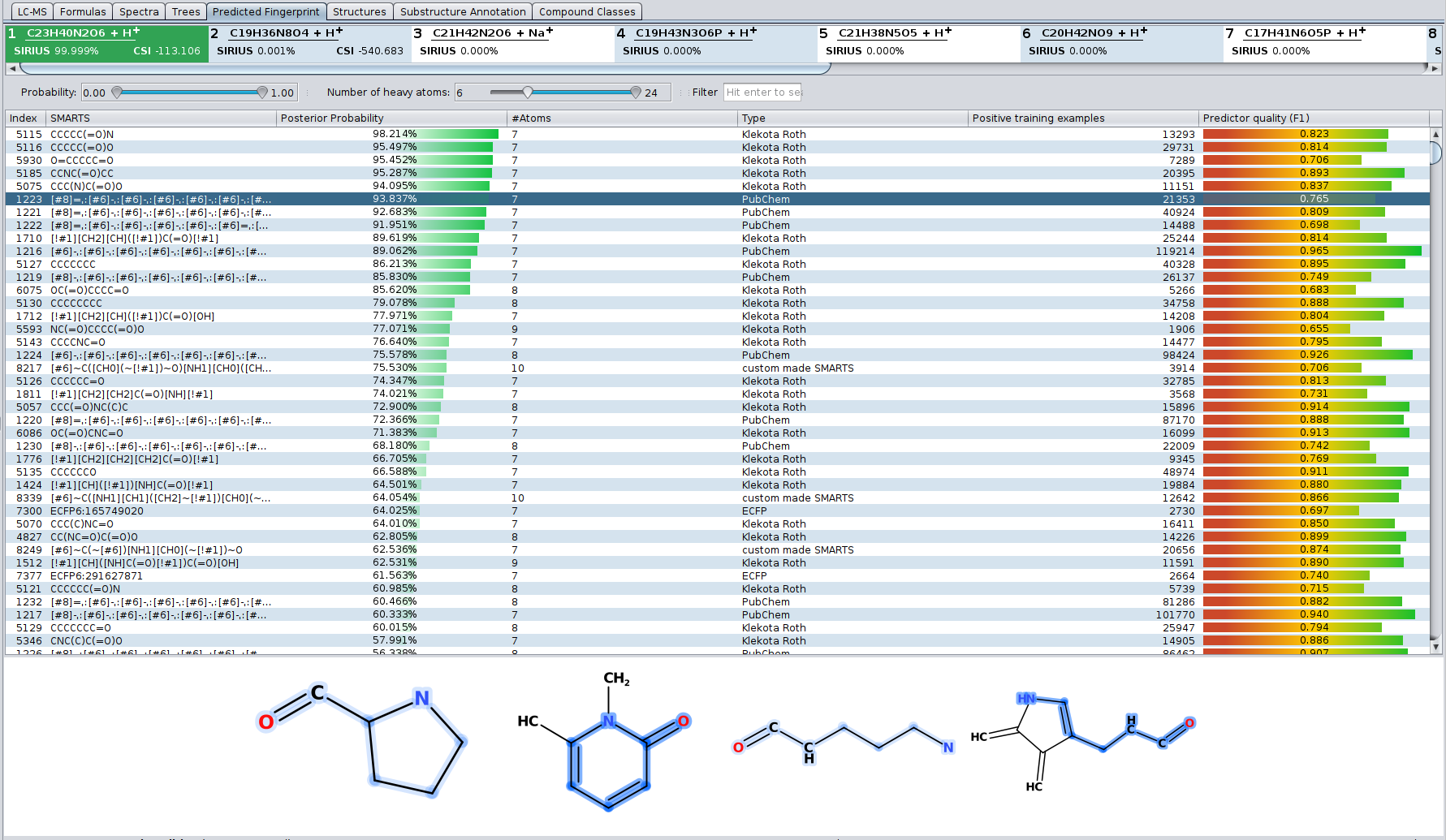
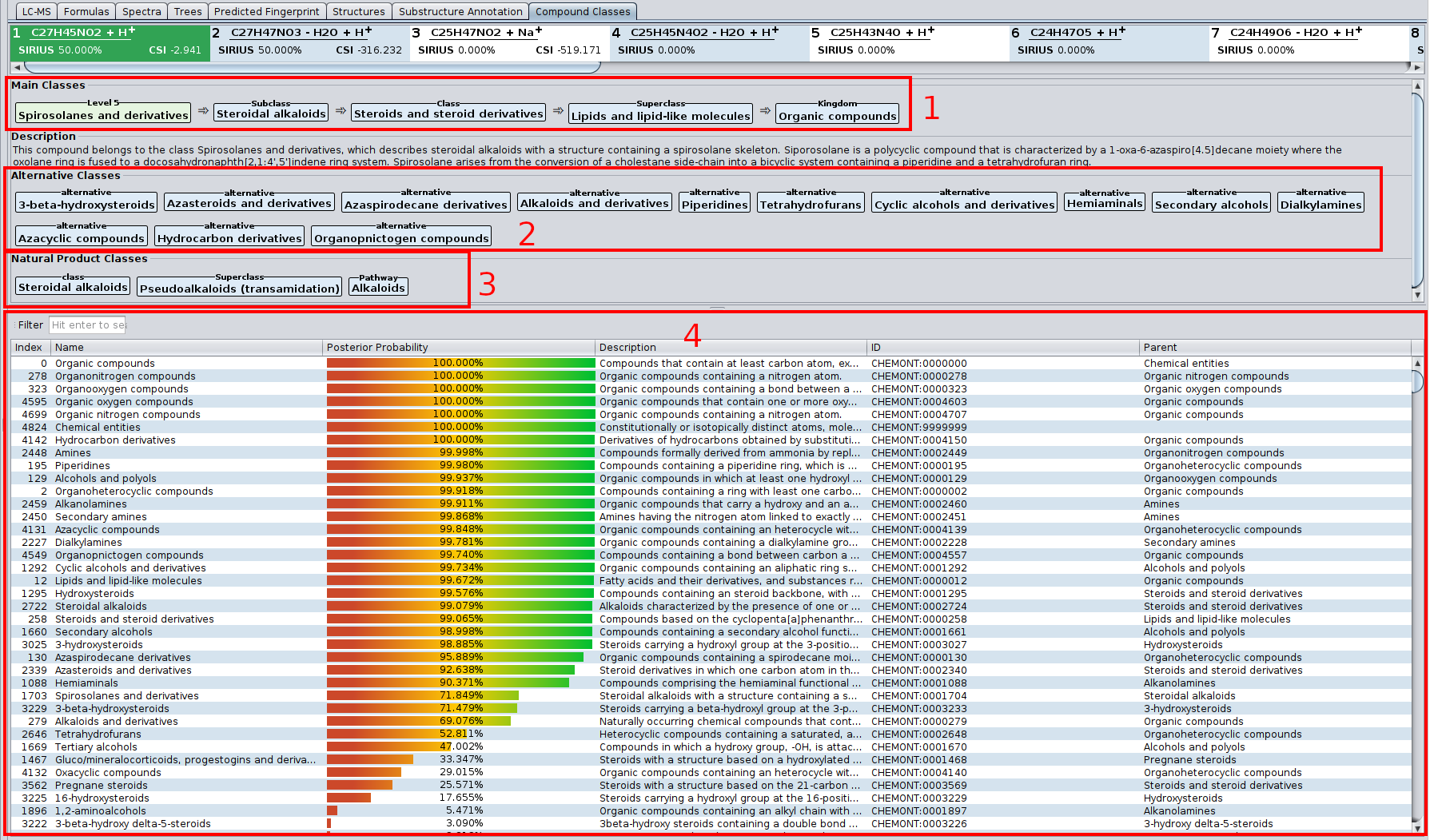
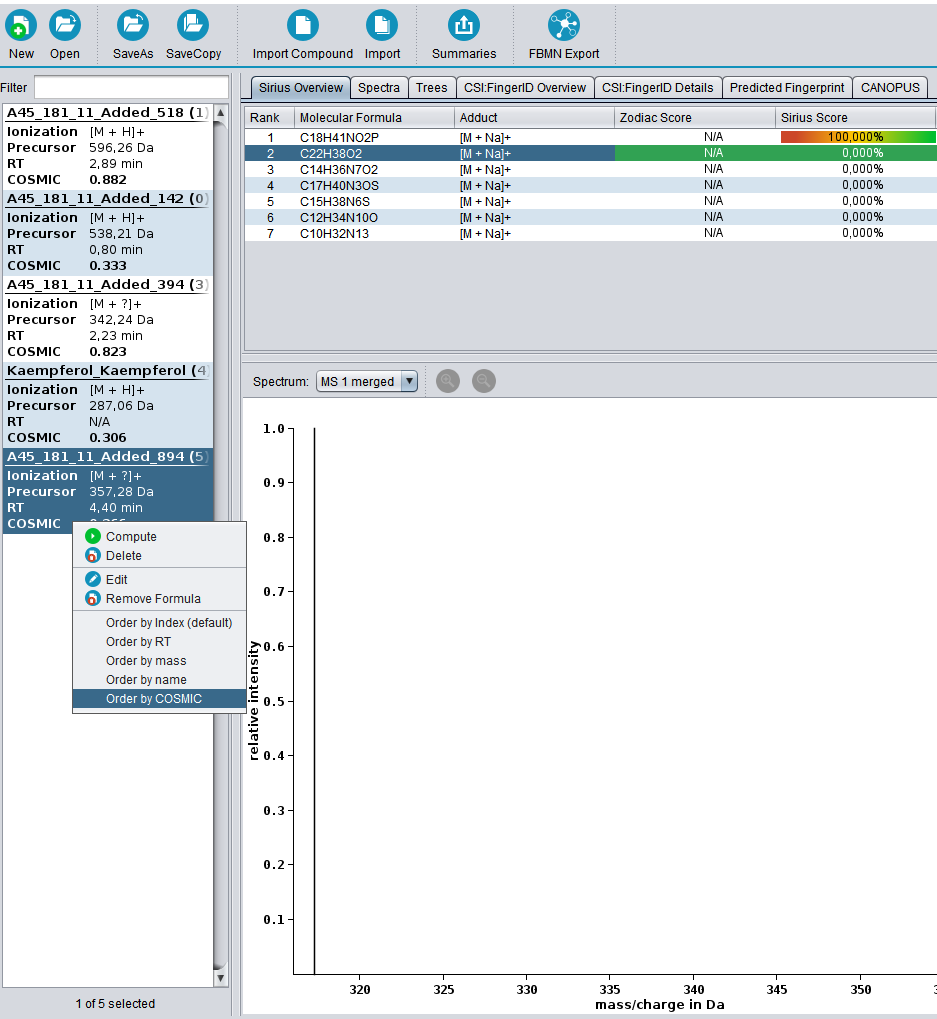
? Improved Result Views: Numerous enhancements and fixes across result views for a smoother analysis experience.