On June 24, Sebastian will give a presentation on SIRIUS 6 at the Okinawa Workshop for Computational Mass Spectrometry.
News
Meet Jonas at the Computational Metabolomics Workshop in Aberdeen
At the Computational Mass Spectrometry Workshop (19-21 June 2024), Jonas will hold a presentation on SIRIUS 6.
Meet Sebastian, Kai and Fleming at the Metabolomics conference in Osaka
At the Metabolomics 2024 (16-20 June), Sebastian will give a keynote presentation. Kai and Fleming will hold a workshop on SIRIUS 6.
Meet Markus at the ASMS conference in Anaheim
Meet Markus at the ASMS conference 2024 in Anaheim between June 2-6!
Meet Andrés and Nils at the European School of Metabolomics in Granada
Meet us at the EUSM 2024 in Granada! Nils will give a hands-on session on SIRIUS.
Prague workshop will be streamed
Good news for those who want to attend the Prague workshop but were not admitted: The Prague workshop on computational mass spectrometry will be streamed, see here. You should get the necessary software installed beforehand if you want to join in.
IMPRS call for PhD student
The International Max Planck Research School at the Max Planck Institute for Chemical Ecology in Jena is looking for PhD students. One of the projects (Project 7) is from our group on “rethinking molecular networks”. Application deadline is April 19, 2024.
Mass spectrometry (MS) is the analytical platforms of choice for high-throughput screening of small molecules and untargeted metabolomics. Molecular networks were introduced in 2012 by the group of Pieter Dorrestein, and have found widespread application in untargeted metabolomics, natural products research and related areas. Molecular networking is basically a method of visualizing your data, based on the observation that similar tandem mass spectra (MS/MS) often correspond to compounds that are structurally similar. Constructing a molecular network allows us to propagate annotations through the network, and to annotate compounds for which no reference MS/MS data are available. Since its introduction, the computational method has received a few “updates”, including Feature-Based Molecular Networks and Ion Identity Molecular Networks. Yet, the fundamental idea of using the modified cosine to compare tandem mass spectra, has basically remained unchanged at the core of the method.
In this project, we want to “rethink molecular networks”, replacing the modified cosine by other measures of similarity, including fragmentation tree similarity, the Tanimoto similarity of the predicted fingerprints, and False Discovery Rate estimates. See the project description for details.
We are searching for a qualified and motivated candidate from bioinformatics, machine learning, cheminformatics and/or computer science who want to work in this exciting, quickly evolving interdisciplinary field. Please contact Sebastian Böcker in case of questions. Payment is 0.65 positions TV-L E13.
IMPRS: https://www.ice.mpg.de/129170/imprs
MPI-CE: https://www.ice.mpg.de/
SIRIUS & CSI:FingerID: https://bio.informatik.uni-jena.de/software/sirius/
Literature: https://bio.informatik.uni-jena.de/publications/ and https://bio.informatik.uni-jena.de/textbook-algoms/
Jena is a beautiful city and wine is grown in the region:
https://www.youtube.com/watch?v=DQPafhqkabc
https://www.google.com/search?q=jena&tbm=isch
https://www.study-in.de/en/discover-germany/german-cities/jena_26976.php
Prague workshop on computational MS overbooked
Unfortunately, the Prague Workshop on Computational Mass Spectrometry (April 15-17, 2024) is heavily overbooked. The organizers will try to stream the workshop and the recorded sessions will be made available online, so check there regularly.
The workshop is organized by Tomáš Pluskal and Robin Schmid (IOCB Prague). Marcus Ludwig (Bright Giant) and Sebastian will give a tutorial on SIRIUS, CANOPUS etc.
Topics of the workshop are: MZmine, SIRIUS, matchms, MS2Query, LOTUS, GNPS, MassQL, MASST, and Cytoscape.
Meet Sebastian at the DGMS conference in Freising
Meet Sebastian at the conference of the Deutsche Gesellschaft für Massenspektrometrie (DGMS 2024) in Freising! The conference is March 10-13, and Sebastian will give a keynote talk on Tuesday, March 12.
BTW, another keynote will be given by our close collaboration partner Michael Witting, also on March 12.
RepoRT has appeared in Nature Methods
Our paper “RepoRT: a comprehensive repository for small molecule retention times” has just appeared in Nature Methods. This is joint work with Michael Witting (Helmholtz Zentrum München) as part of the DFG project “Transferable retention time prediction for Liquid Chromatography-Mass Spectrometry-based metabolomics“. Congrats to Fleming, Michael and all co-authors! In case you do not have access to the paper, you can find the preprint here and a read-only version here.
RepoRT is a repository for retention times, that can be used for any computational method development towards retention time prediction. RepoRT contains data from diverse reference compounds measured on different columns with different parameters and in different labs. At present, RepoRT contains 373 datasets, 8809 unique compounds, and 88,325 retention time entries measured on 49 different chromatographic columns using varying eluents, flow rates, and temperatures. Access RepoRT here.
If you have measured a dataset with retention times of reference compounds (that is, you know all the compounds identities) then please, contribute! You can either upload it to GitHub yourself, or you can contact us in case you need help. In the near future, a web interface will become available that will make uploading data easier. There are a lot of data in RepoRT already, but don’t let that fool you; to reach a transferable prediction of retention time and order (see below), this can only be the start.
If you want to use RepoRT for machine learning and retention time or order prediction: We have done our best to curate RepoRT: We have searched and appended missing data and metadata; we have standardized data formats; we provide metadata in a form that is accessible to machine learning; etc. For example, we provide real-valued parameters (Tanaka, HSM) to describe the different column models, in a way that allows machine learning to transfer between different columns. Yet, be careful, as not all data are available for all compounds or datasets. For example, it is not possible to provide Tanaka parameters for all columns; please see the preprint on how you can work your way around this issue. Similarly, not all compounds that should have an isomeric SMILES, do have an isomeric SMILES; see again the preprint. If you observe any issues, please let us know. See this interesting blog post and this paper as well as our own preprint on why providing “clean data” as well as “good coverage” are so important issues for small molecule machine learning.
Bioinformatische Methoden in der Genomforschung muss leider ausfallen
Nach aktuellem Kenntnisstand muss das Modul “Bioinformatische Methoden in der Genomforschung” im WS 23/24 leider ausfallen. Wir dürfen die Mitarbeiterstelle nicht besetzen, die wir dafür zwingend brauchen. Wir haben gekämpft und argumentiert und alles getan was wir konnten, aber am Ende war es leider vergeblich. Das Modul findet voraussichtlich das nächste Mal im WS 25/26 statt.
Warnung: Im Zuge der Sparmaßnamen an der FSU Jena kann es in Zukunft häufiger zu sollen kurzfristigen Ausfällen kommen.
Meet Sebastian at the Munich Metabolomics Meeting
Sebastian will give a tutorial on using SIRIUS and beyond at the Munich Metabolomics Meeting 2023.
HUMAN EU PhD position is still open
Unfortunately, we have not been able to fill the PhD position for the HUMAN EU project so far. In case you are interested, please contact us!
Update: The position has been filled.
Neues Video zum Studium Bioinformatik
Im Rahmen des MINT Festivals in Jena hat Sebastian ein neues Video zum Studium der Bioinformatik aufgenommen: “Kleine Moleküle. Was uns tötet, was uns heilt“. Es richtet sich vom Vorwissen her an Schüler aus der Oberstufe, aber vielleicht können auch Schüler aus den Jahrgangsstufen darunter etwas mitnehmen. Das Video ist erst mal nur über diese Webseite zu erreichen, wird aber in Kürze bei YouTube hochgeladen.
Am Ende noch zwei Fragen: Erstens, welche Zelltypen im menschlichen Körper enthalten nicht die (ganze) DNA? Da war ich aus Zeitgründen bewusst schlampert; keine Regel in der Biologie ohne Ausnahme. Und zweitens, NMR Instumente werden häufig nicht mit flüssigem Stickstoff gekühlt, sondern mit… was? Wie so oft geht es dabei ums liebe Geld.
Meet Nils, Wei, Fleming and Sebastian at GCB 2023
Meet us at the GCB 2023 in Hamburg! Nils, Wei and Fleming are going to present posters, and Sebastian will give a keynote talk.
Meet Sebastian (remotely) and Fleming at the Swedish TB meeting
Sebastian and Fleming will participate in the Swedish National Tuberculosis meeting: Sebastian will give a talk remotely and Fleming will be on site in Umeå to give a hands-on session on SIRIUS.

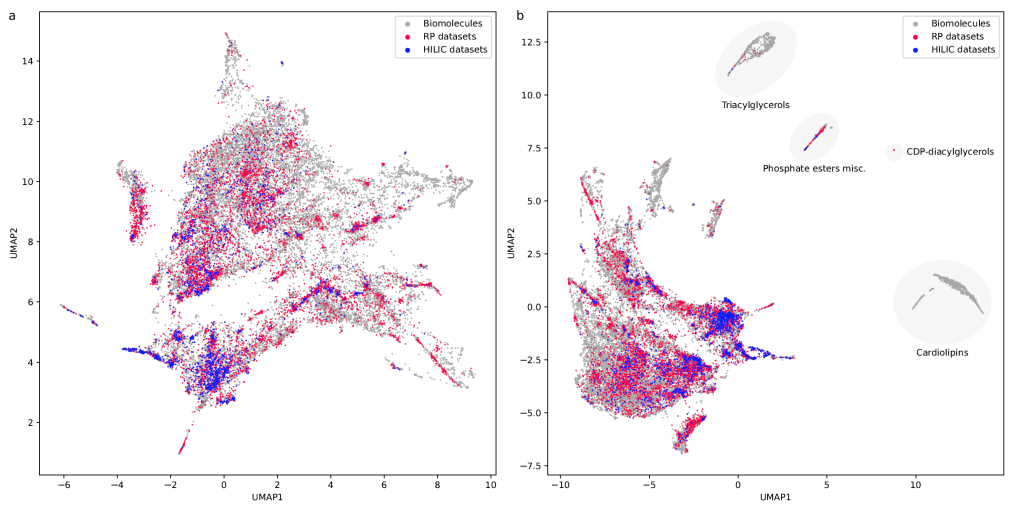
Visualizing the universe of small biomolecules
Have you ever wanted to look at the universe of biomolecules (small molecules of biological interest, including metabolites and toxins)? Have you ever wondered how your own dataset fits into this universe? In our preprint, we introduce a method to do just that, using MCES distances to create a UMAP visualization. Onto this visualization, any compound-dataset can be projected, see the interactive example below. In case it is slow, download the code here. Move your mouse over any dot to see the underlying molecular structure.
If you are wondering, “where did the lipids go?”, check this out. See the preprint on why we excluded them above. Looking at commonly used datasets for small molecule machine learning, big differences can be seen in the coverage of the biomolecule space. For example, the toxicity datasets Tox21 and ToxCast appear to rather uniformly cover the universe of biomolecules. In contrast, SMRT is a massive retention time dataset, but appears to be concentrated on a specific area of the compound space. The thing is: One must not expect a machine learning model trained on only a small part of the “universe of biomolecules”, to be applicable to the whole universe. This is a little too much to be asked. Hence, visualizing your data in this way may give you a better understanding of what your machine learning model is actually doing, where it will thrive and where it might fail.
To compare molecular structures, we compute the MCES (Maximum Common Edge Subgraph) of the two molecular structures. Doing so is not new, but comes at the prize that computing a single distance is already an NP-hard problem, see below. Then, why on Earth are we not using Tanimoto coefficients computed from molecular fingerprints, just like everybody else does? Tanimoto coefficients and related fingerprint-based similarity and dissimilarity measures have a massive advantage over all other means of comparing molecular structures: As soon as you have computed the fingerprints of all molecular structures, computing Tanimoto coefficients is blindingly fast. Hence, if you are querying a database, molecular fingerprints are likely the method of choice. We ourselves have been and are heavily relying on molecular fingerprints: CSI:FingerID is predicting molecular fingerprints from MS/MS data, CANOPUS is predicting compound classes from molecular fingerprints, and COSMIC is using Tanimoto coefficients because they are, well, fast. Yet, if you have ever worked with molecular fingerprints and, in particular, Tanimoto coefficients yourself, you must have also noticed their peculiarities, quirks and shortcomings. In fact, from the moment people used Tanimoto coefficients, others have warned about these unexpected and highly undesirable behaviors; an early example is by Flower (1998). On the one hand, a Tanimoto coefficient of below 0.7 can be the result of two compounds with only one hydroxy group added. On the other hand, two highly different compounds, one half the size of the other, may also have a Tanimoto coefficient of 0.7. Look at the two examples below: According to the Tanimoto coefficient, the two structures on the left are less similar than the two on the right. Does that sound right? By the way: The same holds true for any fingerprint-based similarity or dissimilarity measure, and also for any other fingerprint type. These are examples but the problem is universal.
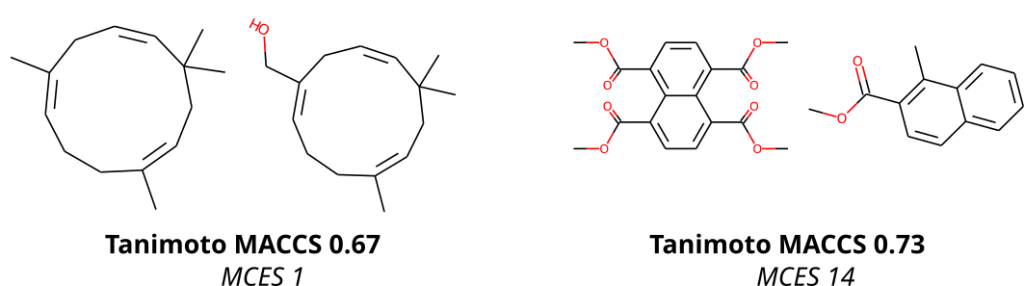
In contrast, the MCES distance is much more intuitive to interpret, as it is the edit distance between molecule graphs and, hence, nicely represents our intuition of chemical reactions. For example, adding an hydroxy group results in an MCES distance of one. Don’t get us wrong: The MCES distance is not perfect, either. First and foremost, the MCES problem is NP-hard; hence, computing a single exact distance between two molecules might take days or weeks. We can happily report that we have “solved” this issue by introducing the myopic MCES distance: We first quickly compute a lower bound on the true distance. If this bound tells us that the true distance is larger than 22, then we would argue that knowing the exact value (maybe 22, maybe 25, maybe 32) is of little help: These two molecules are very, very different, full stop. But if we find that the lower bound only guarantees that the distance is small (say, at most 20) then we use exact computations based on solving an Integer Linear Program. With some more algorithm engineering, we were able to bring down computation time to fractions of a second. And that means that we were able to compute all distances for a set of 20k biomolecular structures, plus several well-known machine learning datasets, in reasonable time and on our limited compute resources. (Sadly, we still do not own a supercomputer.) You will not be able to do all-against-all with a million molecular structures, so if your research requires to do so, you might have to stick with the Tanimoto coefficient, quirky as it is. Yet, we found that subsampling does indeed give us rather reproducible results, see Fig. 7 of the preprint (page 23).
There are other shortcomings of the MCES distance: For example, it is not well-suited to capture the fact that one molecular structure is a substructure of the other. This is unquestioned, but what is also true, is: The MCES distance does not have peculiarities or quirks. The fact that it does not capture substructures, can be readily derived from its definition; this behavior is by design. In case you do not like the absolute MCES distance, because you think that large molecules are treated unfairly, then feel free to normalize it using the size of the molecules. Now that we can (relatively) swiftly compute the myopic MCES distance, we can play around with it.
We used UMAP (Uniform manifold approximation and projection) to visualize the universe of biomolecules but, honestly, we don’t care. You prefer t-SNE? Use that! You prefer a tree-based visualization? Use that! See the following comparison (from left to right UMAP, t-SNE and a Minimum Spanning Tree), created in just a few minutes. Or, maybe Topological Data Analysis? Fine, too! All those visualizations have their pros and cons, and one should always keep Pachter’s elephant in the back of one’s brain. But the thing is: We know that the space of molecular structures has an intrinsic structure, and we are merely using the different visualization methods to get a feeling for its intrinsic structure.
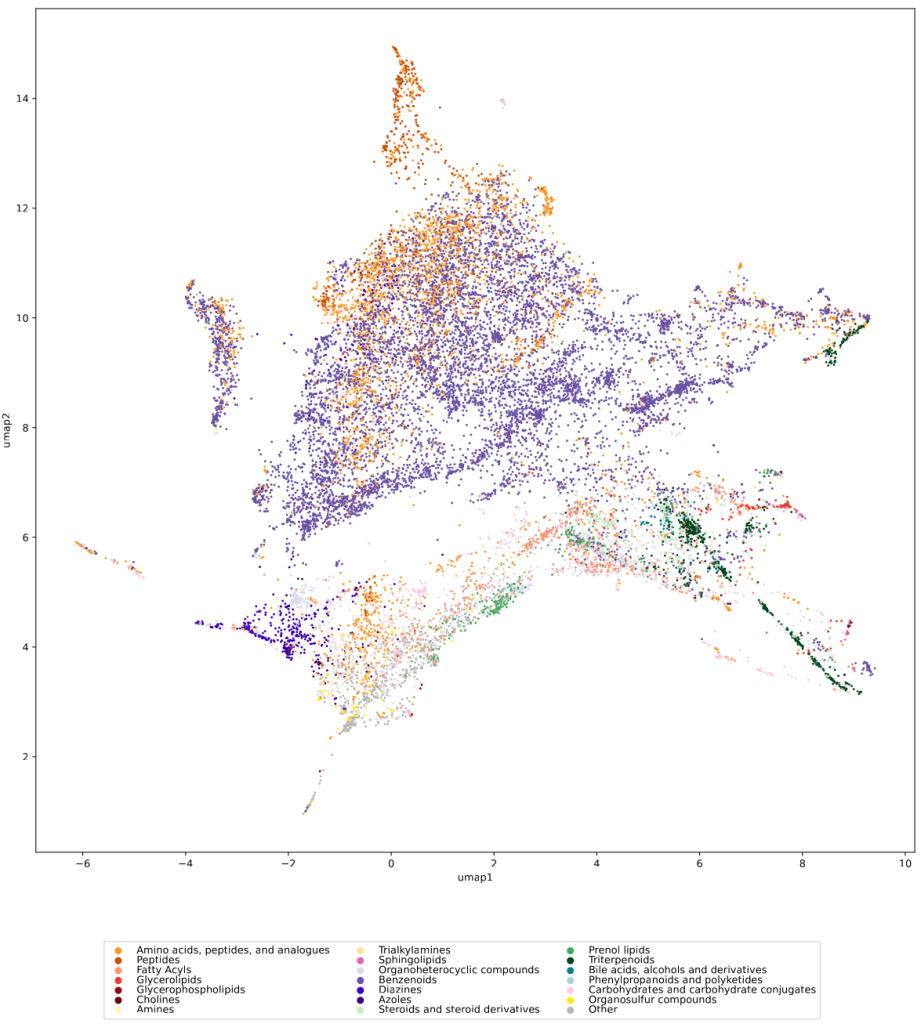
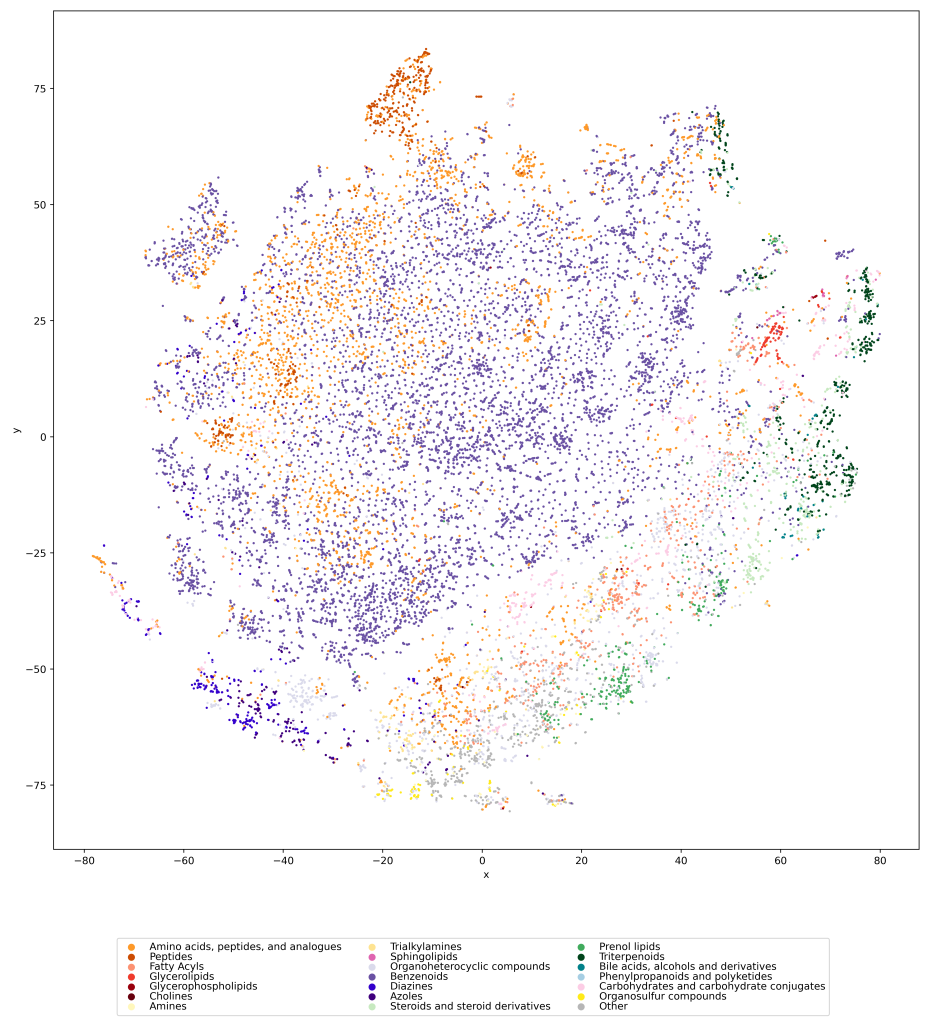

Now, one peculiarity of the above UMAP plots must be mentioned here: When comparing different ML training datasets, we re-used the UMAP embedding computed from the biomolecular structures alone (Fig. 3 in the preprint). Yet, UMAP will neatly integrate any new structures into the existing “compound universe” even if those new structures are very, very different from the ones that were used to compute the projection. This is by design of UMAP, it interpolates, all good. So, we were left with two options: Recompute the embedding for every subplot? This would allow us to spot if a dataset contains compounds very different from all biomolecular structures, but would result in a “big mess” and an uneven presentation. Or, should we keep the embedding fixed? This makes a nicer plot but hides “alien compounds”. We went with the second option solely because the overall plot “looks nicer”; in practice, we strongly suggest to also compute a new UMAP embedding.
In the preprint, we discuss two more ways to check whether a training dataset provides uniform coverage of the biological compound space: We examine the compound class distribution, to check whether certain compound classes are “missing” in our training dataset. And finally, we use the Natural Product-likeness score distribution to check for lopsidedness. All of that can give you ideas about the data you are working with. There have been numerous scandals about machine learning models repeating prejudice in the training data; don’t let the distribution of molecules in your training data let you draw conclusions which, at closer inspection, might be lopsided or even wrong.
If you want to compute myopic MCES distances yourself, find the source code here. You will need a cluster node, or proper patience if you do computations on your laptop. All precomputed myopic MCES distances from the preprint can be found here. We may also be able to help you with further computations.
Retention time repository preprint out now
The RepoRT (well, Repository for Retention Times, you guessed it) preprint is available now. It has been a massive undertaking to get to this point; honestly, we did not expect it to be this much work. It is about diverse reference compounds measured on different columns with different parameters and in different labs. At present, RepoRT contains 373 datasets, 8809 unique compounds, and 88,325 retention time entries measured on 49 different chromatographic columns using varying eluents, flow rates, and temperatures. Access RepoRT here.
If you have measured a dataset with retention times of reference compounds (that is, you know all the compounds identities) then please, contribute! You can either upload it to GitHub yourself, or you can contact us in case you need help. In the near future, a web interface will become available that will make uploading data easier. There are a lot of data in RepoRT already, but don’t let that fool you; to reach a transferable prediction of retention time and order (see below), this can only be the start.
If you want to do anything with the data, be our guests! It is available under the Creative Commons License CC-BY-SA.
If you want to use RepoRT for machine learning and retention time or order prediction, then, perfect! That is what we intended it for. 🙂 We have done our best to curate RepoRT: We have searched and appended missing data and metadata; we have standardized data formats; we provide metadata in a form that is accessible to machine learning; etc. For example, we provide real-valued parameters (Tanaka, HSM) to describe the different column models, in a way that allows machine learning to transfer between different columns. Yet, be careful, as not all data are available for all compounds or datasets. For example, it is not possible to provide Tanaka parameters for all columns; please see the preprint on how you can work your way around this issue. Similarly, not all compounds that should have an isomeric SMILES, do have an isomeric SMILES; see again the preprint. If you observe any issues, please let us know.
Most wanted: Tanaka and HSM parameters for RP columns
A few years ago, Michael Witting and I joined forces to get a transferable prediction of retention times going: That is, we want to predict retention times (more precisely, retention order) for a column even if we have no training data for that column. Yet, to describe a column to a machine learning model, you have to provide some numerical values that allow the model to learn what columns are similar, and how similar. We are currently focusing on reversed-phase (RP) columns because there are more datasets available, and also because it appears to be much easier to predict retention times for RP.
Tanaka parameters and Hydrophobic Subtraction Model (HSM) parameters are reasonable choices for describing a column. Unfortunately, for many columns that are in “heavy use” by the metabolomics and lipidomics community, we do not know these parameters! Michael recently tweeted about this problem, and we got some helpful literature references — kudos! for that. Yet, there are still many columns in the unknown.
Now, the problem is not so much that the machine learning community will not be able to make use of training data from these columns, simply because a few column parameters are unknown. This is unfortunate, but so be it. The much bigger problem is that even if someone comes up with a fantastic machine learning model for transferable retention time prediction — it may not be applicable for your column. Because for your column we do not know the parameters! That would be very sad.
So, here is a list of columns that are heavily used, but where we do not know Tanaka parameters, HSM parameters, or both. Columns are ordered by “importance to the community”, whatever that means… If you happen to know parameters for any of the columns below, please let us know! You can post a comment below or write us an email or send a carrier pigeon, whatever you prefer. Edit: I have switched off comments, it was all spam.
Missing HSM parameters
- Waters ACQUITY UPLC HSS T3
- Waters ACQUITY UPLC HSS C18
- Restek Raptor Biphenyl
- Waters CORTECS UPLC C18
- Phenomenex Kinetex PS C18
Missing Tanaka parameters
- Waters CORTECS T3
- Waters ACQUITY UPLC HSS T3
- Waters ACQUITY UPLC HSS C18
- Restek Raptor Biphenyl
- Waters CORTECS UPLC C18
- Phenomenex Kinetex PS C18