After 2 and a half successful years and over 35 million predicted fingerprints, SIRIUS 4.0.1 will reach its end of life on Friday the 30th of April 2021.
What does this mean for you? We will shut down the web service for CSI:FingerID, so no fingerprint prediction and structure database search will be possible with SIRIUS 4.0.1 anymore.
SIRIUS 4.7.0 Released
We are happy to announce that SIRIUS 4.7.0 is now available for download . This release is all about fixing bugs and performance optimization. To all who had problems with the ILP solvers, a freezing GUI, high memory consumption or long running times: This update should make your life way easier. For a full list of changes see the Changelog.
We further integrated the option to compute fragmentation trees only with our heuristic algorithm (no ILP involved) to speedup molecular formula identification for high mass compounds.
Together with applying timeouts on compound level this should make the processing of large datasets much more feasible.
Introducing COSMIC to assign confidence to annotations
We are happy to introduce COSMIC, a tool for that allows you to assign confidence to structure annotations. For every structure annotated by CSI:FingerID, COSMIC provides a confidence score (a number between 0 and 1) that tells you how likely it is that this annotation is correct. This is similar in spirit to what is done in spectral library search: Not only is the cosine score used to decide which candidate best fits to the query spectrum; in addition, we use the cosine of the top-scoring candidate (the hit) to decide whether it is likely correct (say, above 0.8), incorrect (say, below 0.6) or in the “twilight” in-between. If you have been using CSI:FingerID for some time, you might have noticed that finding such thresholds is not possible for the CSI:FingerID score. COSMIC closes this gap and tells you if an annotation is likely correct or incorrect.
Deciding whether a certain CSI:FingerID hit is correct or incorrect, is undoubtedly convenient in practice. But this is not what COSMIC is all about. What we can do now is to sort all hits in a dataset or even a repository with respect to confidence, and then concentrate our downstream analysis on high-confidence annotations. Next, we can replace the “usual” structure databases we search in by a structure database made entirely from hypothetical structures generated by combinatorics, machine learning or in silico enzymatic reactions.
We demonstrate COSMIC’s power by generating a database of hypothetical bile acid structures, combinatorially adding amino acids to bile acid cores, yielding 28,630 plausible bile acid conjugate structures. We then searched query MS/MS data from a mice fecal dataset in this combinatorial database, and used the COSMIC confidence score to distinguish between hits that are likely correct or incorrect. We manually evaluated the top 12 hits and found that 11 annotation (91.6%) were likely correct; two annotations were further confirmed using synthetic standards. All 11 bile acid conjugates are “truly novel”, meaning that we could not find those structures in PubChem or any other structure database (or publication). Whereas reporting 11 novel bile acid conjugates may appear rather cool, we argue it is even cooler that we did this without a biological hypothesis beyond “there might be bile acid conjugates out there which nobody knows about”; and that COSMIC found the top bile acid conjugate annotations in a fully automated manner.
We have also annotated 2,666 LC-MS/MS runs from human samples with molecular structures which are currently absent from HMDB, and for which no MS/MS reference data are available; and finally, 17,414 LC-MS/MS runs with annotations for which no MS/MS reference data are available. We hope that some of them might be of interest to you.
See the COSMIC preprint for details, and the COSMIC web page for further information. Update: COSMIC is now available through SIRIUS 4.8 and above, download here.
Video Behind the Scenes: CSI:FingerID
There is a new video available and it is finally explaining CSI:FingerID in much detail — possibly too much detail, the video is more than 2 hours. Covers everything from general thoughts and considerations about in silico methods and methods evaluation, to the details of molecular fingerprints, FingerID and, finally, CSI:FingerID. I am sorry for the bad audio quality, still using my build-in laptop mic.
SIRIUS online documentation now available!
We are happy to announce that the new online documentation for SIRIUS is now available at https://boecker-lab.github.io/docs.sirius.github.io/.
The content is completely written in Markdown which makes contributions by the community very easy. No programming skills required!
Help us with your contributions to make this documentation more comprehensive and useful for the community. See our GitHub repository for
detailed information on how to contribute.
Video tutorials for SIRIUS and beyond
We have started to collect video tutorials that explain how to use our software (thanks to Martin) or that allow you to look behind the scenes. We will continue to add videos, so check for updates.
Classes for the masses: CANOPUS has appeared in Nature Biotechnology
Our article “Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra” has just appeared in Nature Biotechnology. Congrats to Kai and all co-authors!
In short: CANOPUS is a computational tool for systematic compound class annotation. It uses a deep neural network to predict 2,497 compound classes from fragmentation spectra, including all biologically relevant classes. From the machine learning perspective, the interesting part is that different levels of the neural network are trained using different data (heterogeneous training). CANOPUS explicitly targets compounds for which neither spectral nor structural reference data are available, and even predicts classes completely lacking tandem mass spectrometry training data. In evaluation using reference data, CANOPUS reached very high prediction performance (average accuracy of 99.7% in cross-validation) and outperformed four (rather advanced) baseline methods. We used CANOPUS to investigating the effect of microbial colonization in the mouse digestive system, for analyzing the chemodiversity of different Euphorbia plants, and for the structural elucidation of a novel marine natural product.
CANOPUS is already available to users through SIRIUS 4.5, which was released last Thursday. See also the designated CANOPUS page. A view-only version of the article is available here.
Full citation: K. Dührkop, L.-F. Nothias, M. Fleischauer, R. Reher, M. Ludwig, M. A. Hoffmann, D. Petras, W. H. Gerwick, J. Rousu, P. C. Dorrestein, and S. Böcker. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat Biotechnol, 2020. https://doi.org/10.1038/s41587-020-0740-8
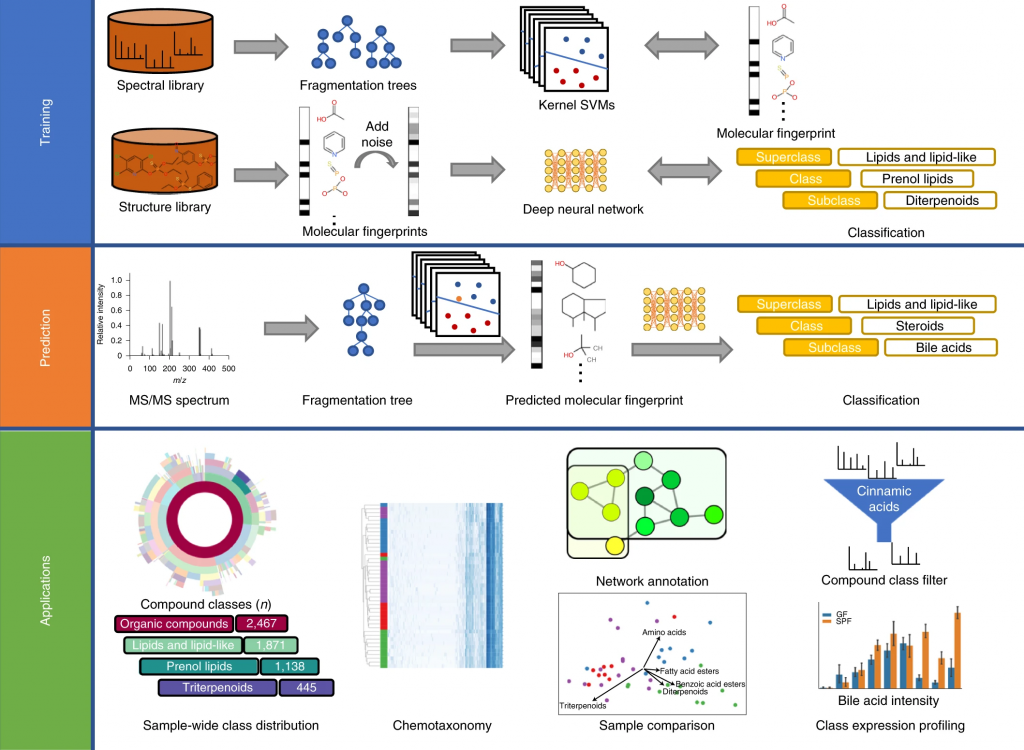
SIRIUS 4.5 released
We are happy to announce that a new version of SIRIUS is available. With that, CANOPUS now supports negative ion mode data. Additionally, we included more structure databases CSI:FingerID can search in, such as COCONUT (Sorokina & Steinbeck, 2020) and NORMAN (Brack et al., 2012). And in case an important database is missing: With the new version, you can import custom databases using the GUI.
Even more features:
- All molecular structures have been standardized using the PubChem standardization service, to make structures more consistent. This update was already reported for version 4.4 but kept bugging us; it should now be solved for good. The standardization has a (small but measurable) positive impact on CSI:FingerID’s performance. More importantly, you will find fewer cases where CSI:FingerID is doing “something really strange”; this strange behavior was often due to un-standardized structures.
- Breaking news: We renamed a few columns in the SIRIUS project space (see Changelog), to make column names more descriptive. Sorry about that; please make sure your downstream analysis is reading in the right columns.
- CSI:FingerID now uses the molecular formula-specific Bayesian network scoring from our ISMB 2018 publication. Integrating this new score was a huge effort, but again has a positive impact on CSI:FingerID’s performance.
- To allow for a smooth transition, you can continue to use SIRIUS 4.4 and the corresponding CSI:FingerID web service until November the 30th.
- Please help us to make SIRIUS great again: Report bugs using the SIRIUS GitHub repository, or send an email to sirius@uni-jena.de.




Qemistree has appeared in Nature Chemical Biology
Congratulations to Anupriya Tripathi from the group of Pieter Dorrestein: The article “Chemically informed analyses of metabolomics mass spectrometry data with Qemistree” has appeared in Nature Chemical Biology, and we are happy to be part of this research.
In short, Qemistree is a data exploration strategy based on the hierarchical organization of molecular fingerprints predicted from fragmentation spectra. Qemistree allows mass spectrometry data to be represented in the context of sample metadata and chemical ontologies.
Full citation: A. Tripathi, Y. Vázquez-Baeza, J. M. Gauglitz, M. Wang, K. Dührkop, M. Nothias-Esposito, D. D. Acharya, M. Ernst, J. J. J. van der Hooft, Q. Zhu, D. McDonald, A. D. Brejnrod, A. Gonzalez, J. Handelsman, M. Fleischauer, M. Ludwig, S. Böcker, L.-F. Nothias, R. Knight, and P. C. Dorrestein. Chemically informed analyses of metabolomics mass spectrometry data with Qemistree. Nat Chem Biol, 2020.
Lehre im Wintersemester 2020/21
Auch im Wintersemester hat uns Corona noch im Griff; deshalb werden die meisten Lehrveranstaltungen online erfolgen. Hier ein paar Details, was Sie erwartet (Achtung, diese news ist eine sticky note; wir werden sie aktualisieren, wenn es weitere Informationen gibt):
- NEU: Das Seminar Beruf und Karriere (ASQ) findet als Blockveranstaltung in der Woche vom 22. bis 26. März statt. Das Seminar ist online und live (Zoom-Veranstaltung). Details folgen, Anmeldung über Friedolin ist bereits möglich.
- Einführung in die Bioinformatik I/1: Die Vorlesung macht Peter Dittrich; die Video-Dateien werden zum Download bereitgestellt und Sie können sich diese anhören/ansehen, wann es Ihnen passt. Wöchentlich am Dienstag von 10:15 bis 11:45 ist das Tutorium bei Sebastian Böcker; auch das wird online angeboten, ist aber live (Zoom-Veranstaltung). Die beiden Übungen sind parallel wöchentlich am Mittwoch von 14:15 bis 15:45. Das sind Präsenzveranstaltungen, und sie finden im Hörsaal 1 des Abbeanum und im Seminarraum 104 in der August-Bebel-Str. 4 statt. (Edit: Es sieht aktuell so aus, dass wir die Übungen als Präsenzveranstaltungen durchführen können.) Übungsleiter sind Marcus Ludwig und Emanuel Barth. Die Aufteilung auf die beiden Gruppen können wir im ersten Tutorium vornehmen. Die Veranstaltung startet mit dem Tutorium am Dienstag 3. November.
- Algorithmische Massenspektrometrie: VL und Tutorium bei Sebastian Böcker. Die VL wird als Video-Dateien zum Download bereitgestellt, das Tutorium ist online aber live (Zoom-Veranstaltung), wöchentlich am Montag von 12:3o bis 14:oo. Die Übung ist wöchentlich am Donnerstag 12-14 Uhr, online aber live (Zoom-Veranstaltung), Übungsleiter ist Kai Dührkop. Die Veranstaltung startet mit dem Tutorium am Montag 2. November.
- Currents in Bioinformatics: Das Seminar findet wöchentlich Dienstags 16:15 bis 17:45 statt (online aber live, Zoom-Veranstaltung). Ansprechpartner ist Fleming Kretschmer. Regelmäßige Teilnahme ist zwingend erforderlich. Wir lesen und besprechen hier aktuelle Forschungs-Paper. Die Veranstaltung startet am Dienstag 3. November.
- Datamining und Sequenzanalyse: Die Veranstaltung wird von Markus Fleischauer gehalten und findet Mo 10:15-11:45h und Fr 12:30-14:00h statt. Wir werden auch diese Veranstaltung online durchführen, sofern alle Studierenden einen geeigneten Computer zur Durchführung der Programmieraufgaben zur Verfügung haben. Ein Computer ist für das Praktikum geeignet, wenn er diese Anforderungen erfüllt. Alle für das Praktikum benötigte Software ist für Linux, Mac und Windows verfügbar und wird während des Praktikums gemeinsam installiert. Sollte jemand keinen geeigneten Computer organisieren können, bitte umgehend bei Markus Fleischauer melden. Wir werden Zoom verwenden. Der Link wird rechtzeitig mitgeteilt. Alle Informationen zur Veranstaltung werden hier zu finden sein. Start ist am Montag 02. November um 10:15 Uhr.
Bei individuellen Problemen und Fragen wenden Sie sich bitte via Email an Peter Dittrich (Studiengangsverantwortlicher) oder Sebastian Böcker. Sie können auch individuelle Gespräche via Zoom vereinbaren.
FSU-Disclaimer zum Vorlesungsmaterial, insbesondere zu den Videos mit den Vorlesungen:
- In unseren Veranstaltungen und ihrer Aufzeichnung wird ggf. urheberrechtlich geschütztes Material verwandt. Eine Nutzung, etwa durch Verbreitung oder Veröffentlichung dieses Materials, ist untersagt und kann die Geltendmachung von Unterlassungs- und Schadensersatzansprüchen zur Folge haben.
ZODIAC has appeared in Nature Machine Intelligence
Our article “Database-independent molecular formula annotation using Gibbs sampling through ZODIAC” has just appeared in Nature Machine Intelligence. Congrats to Marcus and all co-authors!
In short: Annotating the molecular formula of a small molecule is the first step towards its structural elucidation but remains highly challenging, particularly for “large compounds” above 500 Daltons. ZODIAC is a network-based algorithm for the de novo annotation (no database needed) of molecular formulas, and processes complete experimental LC-MS/MS runs. (No metabolite is an island.) In comparison to SIRIUS, previously best-of-class for this task, ZODIAC reduces the error rate of false annotations roughly to the half. And sometimes, much more…
If you have problems accessing the paper: Here is a read-only version.
ZODIAC is already available to users through SIRIUS 4.4. See also the designated ZODIAC page.
Full citation: M. Ludwig, L.-F. Nothias, K. Dührkop, I. Koester, M. Fleischauer, M.A. Hoffmann, D. Petras, F. Vargas, M. Morsy, L. Aluwihare, P.C. Dorrestein, and S. Böcker. Database-independent molecular formula annotation using Gibbs sampling through ZODIAC. Nat Mach Intell 2:629–641, 2020.
Feature-Based Molecular Networking appeared in Nature Methods
Congratulations to Louis-Félix Nothias from the group of Pieter Dorrestein: The article “Feature-based molecular networking in the GNPS analysis environment” has appeared in Nature Methods, and we are happy to be part of this research. (We are lacking behind a little bit with our news.)
In short, FBMN introduces chromatography separation into the molecular networking workflow of GNPS; features with similar mass (and potentially similar MS/MS) but different retention time are now treated separately. See the article for details.
Full citation: L.-F. Nothias, D. Petras, R. Schmid, K. Dührkop, J. Rainer, A. Sarvepalli, I. Protsyuk, M. Ernst, H. Tsugawa, M. Fleischauer, F. Aicheler, A.A. Aksenov, O. Alka, P.-M. Allard, A. Barsch, X. Cachet, A.M. Caraballo-Rodriguez, R.R. Da Silva, T. Dang, N. Garg, J.M. Gauglitz, A. Gurevich, G. Isaac, A.K. Jarmusch, Z. Kameník, K.B. Kang, N. Kessler, I. Koester, A. Korf, A. Le Gouellec, M. Ludwig, C. Martin H., L.-I. McCall, J. McSayles, S.W. Meyer, H. Mohimani, M. Morsy, O. Moyne, S. Neumann, H. Neuweger, N.H. Nguyen, M. Nothias-Esposito, J. Paolini, V.V. Phelan, T. Pluskal, R.A. Quinn, S. Rogers, B. Shrestha, A. Tripathi, J.J.J. van der Hooft, F. Vargas, K.C. Weldon, M. Witting, H. Yang, Z. Zhang, F. Zubeil, O. Kohlbacher, S. Böcker, T. Alexandrov, N. Bandeira, M. Wang, and P.C. Dorrestein. Feature-based molecular networking in the GNPS analysis environment. Nat Methods 17(9):905–908, 2020.
Marcus has successfully defended his thesis last Tuesday
Congratulations!

SIRIUS 4.4.21 – Can now be used along with 4.0.1
Some of you may have noticed problems where SIRIUS 4.4 GUI did not start without reporting any error.
This might be due to old incompatible configs (.sirius directory) from version 4.0.1. SIRIUS 4.4.21 fixes this problem and now uses a separate config directory (.sirius-4.4). It is now possible to use version 4.0.1 and 4.4.x along on the same system without interfering each other.
SIRIUS 4.4 GUI for MacOS X now available
Since we could fix the deadlocks of the SIRIUS GUI on Mac with build 4.4.18, the SIRIUS 4.4. GUI now also available for MacOS: https://bio.informatik.uni-jena.de/software/sirius/
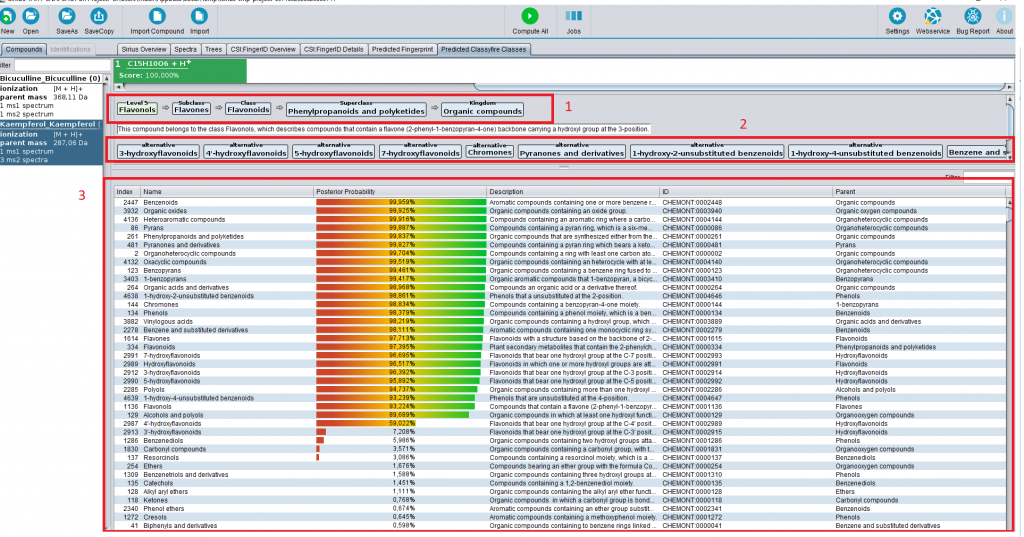
Introducing CANOPUS for comprehensive compound class annotation
We are happy to introduce CANOPUS, a tool for the comprehensive annotation of compound classes from MS/MS data (certain restrictions apply, see below). In principle, CANOPUS is doing something similar as CSI:FingerID: Whereas CSI:FingerID can tell you what substructures are part of the query compound, CANOPUS does so for compound classes. The differences between both tasks are subtle but have massive consequences. See this preprint on the details of this difference, how CANOPUS works, how good it works etc.
At present, CANOPUS predicts 1270 compound classes. In more detail, CANOPUS predicts ClassyFire compound classes. ClassyFire is not the first but, to the best of our knowledge, by far the most comprehensive approach to assign classes solely from structure. (This last point is key, as this allows us to assign thousands of classes for millions of molecular structures.) Please have a look there if you use CANOPUS: Certain compound class definitions may be not what you expect. For example, we found that many phytosteroids are classified as bile acids in ClassyFire. While the biochemical origin of both classes is very different, they are structural very similar and, therefore, represented by the same class in the ClassyFire ontology.
You can download, install and use CANOPUS through SIRIUS 4.4. You will notice a new tab where you can access, for each compound, all compound classes it does or does not belong to (and, how sure we are about that). Fancier visualizations (see the preprint) will be made available with upcoming releases.
ps. Clearly, CANOPUS is comprehensive only within the limits of the LC-MS/MS technology: If a compound does not ionize, if no fragmentation spectrum is recorded in Data Dependent Acquisition, if a compound does not show any fragmentation, if multiple compounds are fragmented in a single spectrum etc, then CANOPUS cannot help you. We don’t do magic. Also, CANOPUS is limited by the available (structure and MS/MS) training data; but several years of thinking have been invested to get the most out of it.
Introducing ZODIAC for improved molecular formula annotations
We are happy to introduce ZODIAC, a tool for the comprehensive annotation of molecular formulas for complete LC-MS/MS runs. SIRIUS 4 is currently best-of-class for this task (as far as we know); but ZODIAC can do better. Different from SIRIUS which considers one compound at a time, ZODIAC considers a complete dataset, assuming that all compounds are somehow related (usually through biotransformations). See the preprint for evaluation and method details.
ZODIAC is about de novo annotations, meaning that we can assign molecular formulas for novel compounds currently absent from any structure database. ZODIAC takes into account “uncommon” elements, as in C24H47BrNO8P or C15H30ClIO5; both examples are indeed novel molecular formulas annotated by ZODIAC (and verified by us). Enter those molecular formulas into the PubChem search and see what you get back. (Fun fact: the first query now returns two entries created Jan 2020 based on our annotations.)
You can download, install and use ZODIAC through SIRIUS 4.4. Results of ZODIAC are simply displayed in the molecular formula tab, if you choose to run it. You should definitely use ZODIAC if you want to run CANOPUS: Assigning molecular classes to novel compounds implies that some of the molecular formulas may be novel, too; and you do not want provide CANOPUS a wrong molecular formula.
ps. Sorry for tweeting early, WordPress sometimes has a mind of its own.
SIRIUS 4.4 released
We are happy to announce that SIRIUS 4.4 is finally released. (Unfortunately, the MacOS version will have to wait a few more days.) There have been numerous changes and improvements, only few of which can be mentioned here.
Probably the biggest change is that SIRIUS 4.4 now reads mzML files (“centroided” data) and processes complete LC-MS/MS datasets. You can use ProteoWizard to transform your dataset to mzML. This does not only make things easier for you; it also allows SIRIUS to extract isotope patterns and adduct information more thoroughly from the MS1 data. SIRIUS 4.4 also supports multi-run datasets and aligns runs.
If you are using the graphical user interface (GUI) you no longer have to care about installing (the correct version of) Java. It is part of the installed SIRIUS software.
SIRIUS 4.4 uses the same project space for the command-line (CLI) and the GUI version, allowing you to use the SIRIUS GUI to browse through results computed with the CLI. The GUI also allows you to save your project and reload it later, including all previously computed results. Finally, you can export summary CSV and mzTab-M files for downstream analysis.
CSI:FingerID also had some updates:
- Additional large molecular substructures: Have a look at the Fingerprint tab in the SIRIUS GUI, filter for large substructures.
- Standardization of molecular structures (mesomerism, charge etc) through PubChem. This does not only improve identification statistics by a few percentage points, but also gets rid of certain cases where CSI:FingerID was doing “strange things”. Unfortunately, PubChem keeps changing the standardization without giving big notice, so some issues remain; but the current situation is definitely better than no standardization.
More stuff:
- There is currently no version for MacOS; we are sorry. Somehow, MacOS does not like our multithreading. At present, we do not have access to a Mac for debugging, thanks to Corona.
- Please report bugs using the SIRIUS GitHub repository or sirius@uni-jena.de. There will be numerous such bugs, as SIRIUS 4.4 again carries major improvements and transformations under the hood. Help us to make SIRIUS better.
- To allow for a smooth transition, you can continue to use SIRIUS 4.0.1 and the corresponding CSI:FingerID web service for a couple of weeks.
- SIRIUS 4.4 integrates ZODIAC and CANOPUS, see the separate news.
- passatutto is integrated into SIRIUS 4.4, allowing you to generate your own spectral library decoy database for FDR estimation.
- We have included a beautiful interactive fragmentation tree viewer.
- There may be a few more releases of SIRIUS 4.4.x that ship those things which are done in principle.
- Finally, we have not reported the number of CSI:FingerID queries for some time, so here we go: There have been 47 million CSI:FingerID queries. (Plus a few million we lost through a little scripting bug. Our bad.) That is roughly one query every 1.5 seconds since we reported one million queries in Feb 2018.
Hiccup of FSU computer network
Yesterday (27 April 2020) our university computer network experienced some issues and was unavailable for several hours. Not unexpectedly, this also resulted in the unavailability of the CSI:FingerID web service, website etc. As usual, computer problems cause more computer problems: It looks like today (28 April 2020) we still have certain issues restarting the CSI:FingerID workers. That is hopefully resolved soon. We apologize for any inconvenience.
SIRIUS 4.4 beta released
Some of you may have noticed that yesterday, April 17, the SIRIUS 4.4 beta has been released. This update is huge so we are particularly careful not to break too many things. (We will definitely break some things so please report bugs using the SIRIUS GitHub repository or sirius@uni-jena.de.) Some facts of what you can expect:
- The official SIRIUS 4.4 release will happen in a few days.
- Even after SIRIUS 4.4 has been officially deployed, you can continue to use SIRIUS 4.0.1 and the corresponding CSI:FingerID web service. We hope that this allows for a smooth transition.
- SIRIUS 4.4 integrates ZODIAC for better molecular formulas.
- SIRIUS 4.4 integrates CANOPUS for compound class assignments.
- SIRIUS 4.4 now reads mzML files (“centroided” data) and processes complete LC-MS/MS datasets.
- CSI:FingerID had some massive updates, including more and larger molecular properties and standardization of molecular structures.
- SIRIUS 4.4 also supports multi-run datasets and aligns runs.
- SIRIUS 4.4 uses the same project space for the command-line and the user interface version, allowing you to use the SIRIUS GUI to browse through results computed with the CLI.
- passatutto is integrated into SIRIUS 4.4, allowing you to generate your own spectral library decoy database for FDR estimation.
- If you wonder why we jump from version 4.0.1 to 4.4: There have been several internal releases in between.
- A word of warning: Many features and changes have accumulated and there will be a few more releases (4.4.x) until the quiver is empty. For example, the structure database will change again as we have massive issues with the way PubChem handles structure standardization.